This is the first of a new format on Genomes Unzipped: as we acquire tests from more companies, or get data from others who have been tested, we’ll post reviews of those tests here. The aim of this series is to help potential genetic testing customers to make an informed decision about the products on the market. We’re still tweaking the format, so if you have any suggestions regarding additional analyses or areas that should be covered in more detail, let us know in the comments.
Overview
Lumigenix is a relative newcomer to the personal genomics scene: the Australian-based company launched back in March this year, offering a SNP chip-based genotyping service similar in concept to those provided by 23andMe, deCODEme and Navigenics.
The company kindly provided Genomes Unzipped with 12 free “Comprehensive” kits, which provide genotypes at over 700,000 positions in the genome, to enable us to review their product. We note that the company offers several other services, including a lower-priced “Introductory” test that covers fewer SNPs, and whole-genome sequencing for the more ambitious personal genomics enthusiast. This review should be regarded as entirely specific to the Comprehensive test.
Continue reading ‘Review of the Lumigenix “Comprehensive” personal genome service’
 I have no strong family history of any disease, despite having 7 blood aunts and uncles and countless cousins. So when I sent my spit off to 23andMe at the start of the Genomes Unzipped project, I was expecting something very similar to
I have no strong family history of any disease, despite having 7 blood aunts and uncles and countless cousins. So when I sent my spit off to 23andMe at the start of the Genomes Unzipped project, I was expecting something very similar to 


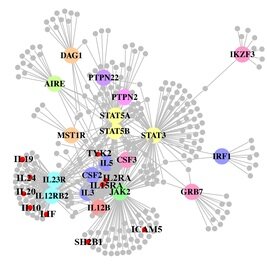 A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
 RSS
RSS Twitter
Twitter
Recent Comments