A lot of the Genomes Unzipped crew seem to be away on holiday at the moment, so today’s Links post may lack the the authorial diversity that you’re accustomed to.
I just got around to reading the August addition of PLoS Genetics, and found a valuable study from the Keck School of Medicine in California. They authors looked at the effect of known common variants in five American ethnic groups (European, African, Hawaiian, Latino and Japanese Americans), to assess how similar or different the effects sizes were across the groups.
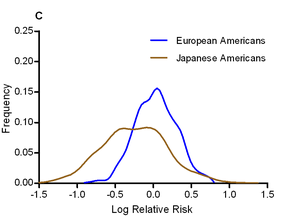 The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
These sorts of datasets will be very useful for personal genomics in the future, as a decade of European-centered genetics research has left non-Europeans somewhat in the lurch with regards to disease risk predictions. However, the problem with the approach in this paper is that even this in large a study (6k cases, 7k controls) the error bounds on the odds ratios within each group are still pretty large. [LJ]
Over at the Guardian Science Blog, Dorothy Bishop explains the difference between learning that a trait is heritable (e.g. from twin studies), and mapping a specific gene “for” a trait (e.g. via GWAS). Her conclusion is worth repeating:
The main message is that we need to be aware of the small effect of most individual genes on human traits. The idea that we can test for a single gene that causes musical talent, optimism or intelligence is just plain wrong. Even where reliable associations are found, they don’t correspond to the kind of major influences that we learned about in school biology. And we need to realise that twin studies, which consider the total effect of a person’s genetic makeup on a trait, often give very different results from molecular studies of individual genes.
There are also interesting questions to be asked about why there is such a gap between heritabilities estimated by twin studies, and the heritability that can be explained by GWAS results. That is, however, is a question for another day. [LJ]
Another article just released in PLoS Genetics provides a powerful illustration of just how routine whole-genome sequencing is now becoming for researchers: the authors report on complete, high-coverage genome sequence data for twenty individuals. The samples included 10 haemophilia patients and 10 controls, taken as part of a larger study looking at the genetic factors underlying resistance to HIV infection. While this is still a small sample size by the standards of modern genomics, there are a few interesting insights that can be gleaned from the data: for instance, the researchers argue from their data that each individual has complete inactivation of 165 protein-coding genes due to genetic variants predicted to disrupt gene function. I’ll be following up on this claim in a future post. [DM]

Finally, a quick shout-out to our fellow Sanger researchers, including Verneri Anttila and Aarno Palotie, along with everyone else in the International Headache Genetics Consortium, for finding the first robust genetic association to migrane. They looked at 3,279 cases and >10k controls (and another 3,202 cases to check their results), and found that the variant rs1835740 was significantly associated with the disease.
To tie in with the above story, in the region of 40-65% of variation in migraine is heritable, but only about 2% of this was explained by the rs1835740 variant. However, explaining heritability isn’t the main point of GWAS studies: a little follow-up found that rs1835740 was correlated with expression of the gene MTDH, which in turn suggests a defect in glutamate transport; hopefully this new discovery will help shed some light on the etiology of the disease. [LJ]
 Over at Nature News, Erika Check Hayden has a post about a recent Science Translational Medicine paper by Bert Vogelstein and colleages looking at the potential predictive power of genetics. The take-home message from the study (or at least the message that has been taken home by, e.g., this NYT article) is that DNA does not perfectly determine which disease or diseases you may get in the future. This take home message is true, and to me relatively obvious (in the same way that smoking doesn’t perfectly determine lung cancer, or body weight and dietary health doesn’t perfectly determine diabetes status).
Over at Nature News, Erika Check Hayden has a post about a recent Science Translational Medicine paper by Bert Vogelstein and colleages looking at the potential predictive power of genetics. The take-home message from the study (or at least the message that has been taken home by, e.g., this NYT article) is that DNA does not perfectly determine which disease or diseases you may get in the future. This take home message is true, and to me relatively obvious (in the same way that smoking doesn’t perfectly determine lung cancer, or body weight and dietary health doesn’t perfectly determine diabetes status). To celebrate 10 years since the back-to-back publications of complete human genomes in Science and Nature, Science has published series of articles looking back at the last 10 years of genomics, and forward to the future. The article contains short essays from Francis Collins and Craig Venter, the former talking about some of the successes of medical sequencing (including giving a name and photograph to the exome-sequenced IBD patient I
To celebrate 10 years since the back-to-back publications of complete human genomes in Science and Nature, Science has published series of articles looking back at the last 10 years of genomics, and forward to the future. The article contains short essays from Francis Collins and Craig Venter, the former talking about some of the successes of medical sequencing (including giving a name and photograph to the exome-sequenced IBD patient I  Of particular interest is an article by Eliot Marshall on why genomics hasn’t yet had a large effect on medical practice, and what needs to be done to allow the genomic revolution to trickle into medical care. He argues that scientists and doctors need to meet each other half way; scientists need to focus more on showing the direct clinical utility of genomics, whereas doctors need to be more ready to accept new technologies and discoveries, and adapt the way they practice medicine to make full use of them. [LJ]
Of particular interest is an article by Eliot Marshall on why genomics hasn’t yet had a large effect on medical practice, and what needs to be done to allow the genomic revolution to trickle into medical care. He argues that scientists and doctors need to meet each other half way; scientists need to focus more on showing the direct clinical utility of genomics, whereas doctors need to be more ready to accept new technologies and discoveries, and adapt the way they practice medicine to make full use of them. [LJ] I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ]
I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ] The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
 RSS
RSS Twitter
Twitter
Recent Comments