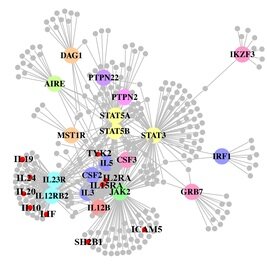
 A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
23andMe customers will be interested in a neat little FireFox plug-in that allows them to view their own genotypes for any 23andMe SNP mentioned on a web page. You can download the plug-in here (you’ll need to have an up-to-date version of FireFox), and I have a brief review of the tool here. [DM]
Continue reading ‘From GWAS to pathways, the consequences of DTC genetics and screening by sequencing’
 RSS
RSS Twitter
Twitter
Recent Comments