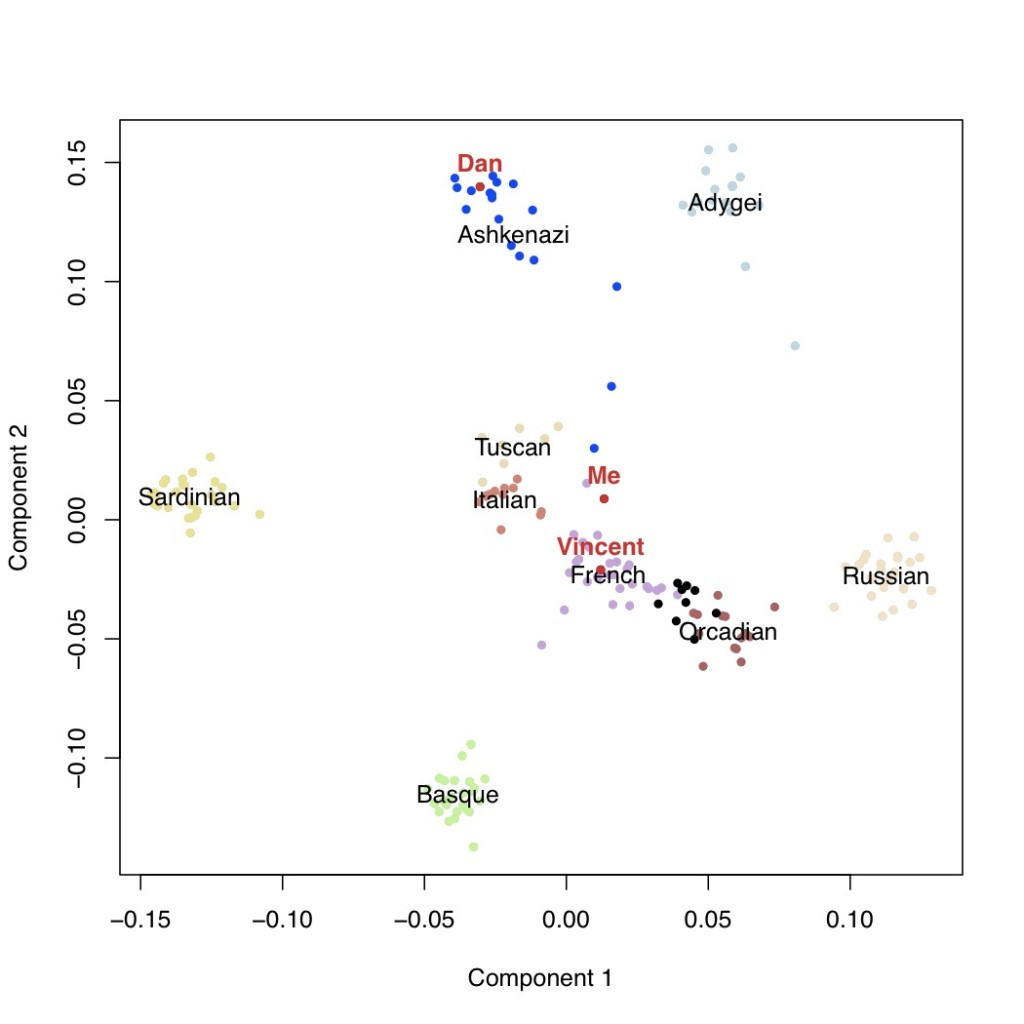
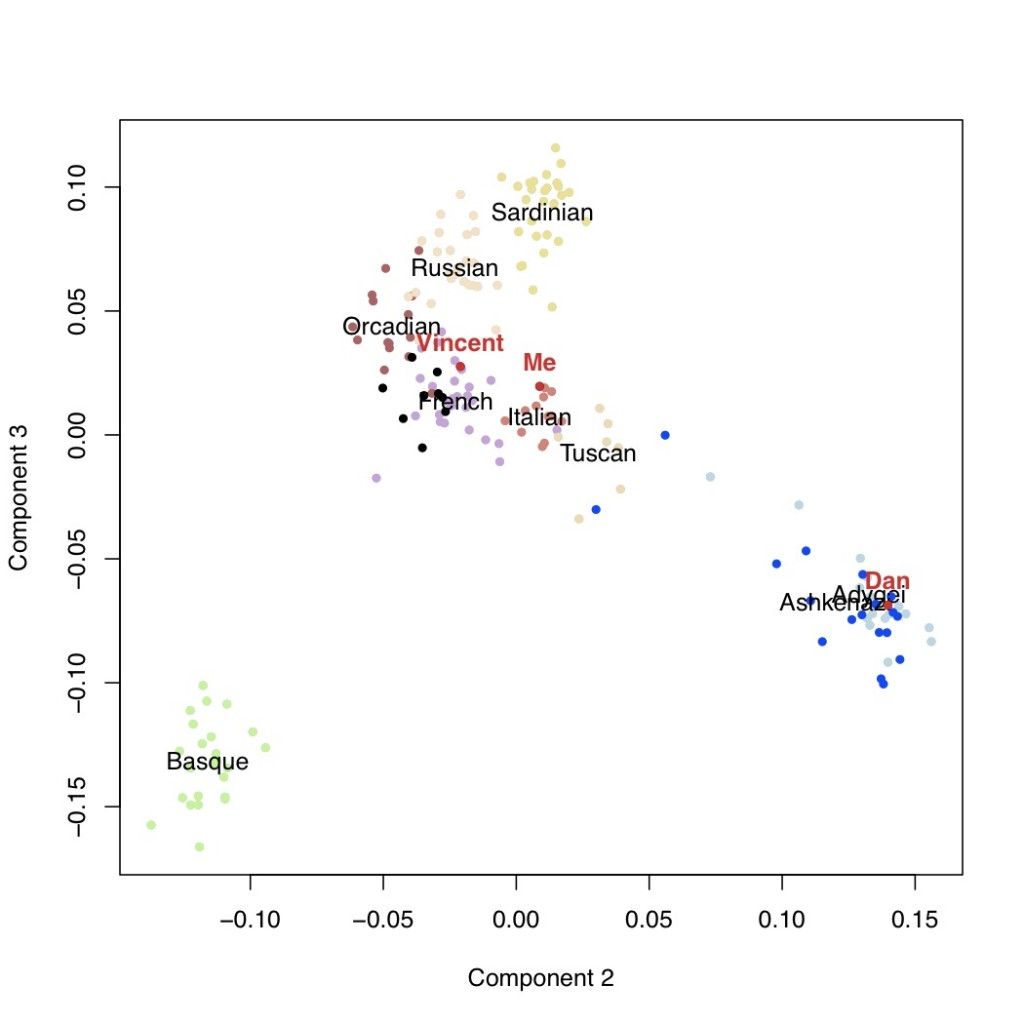
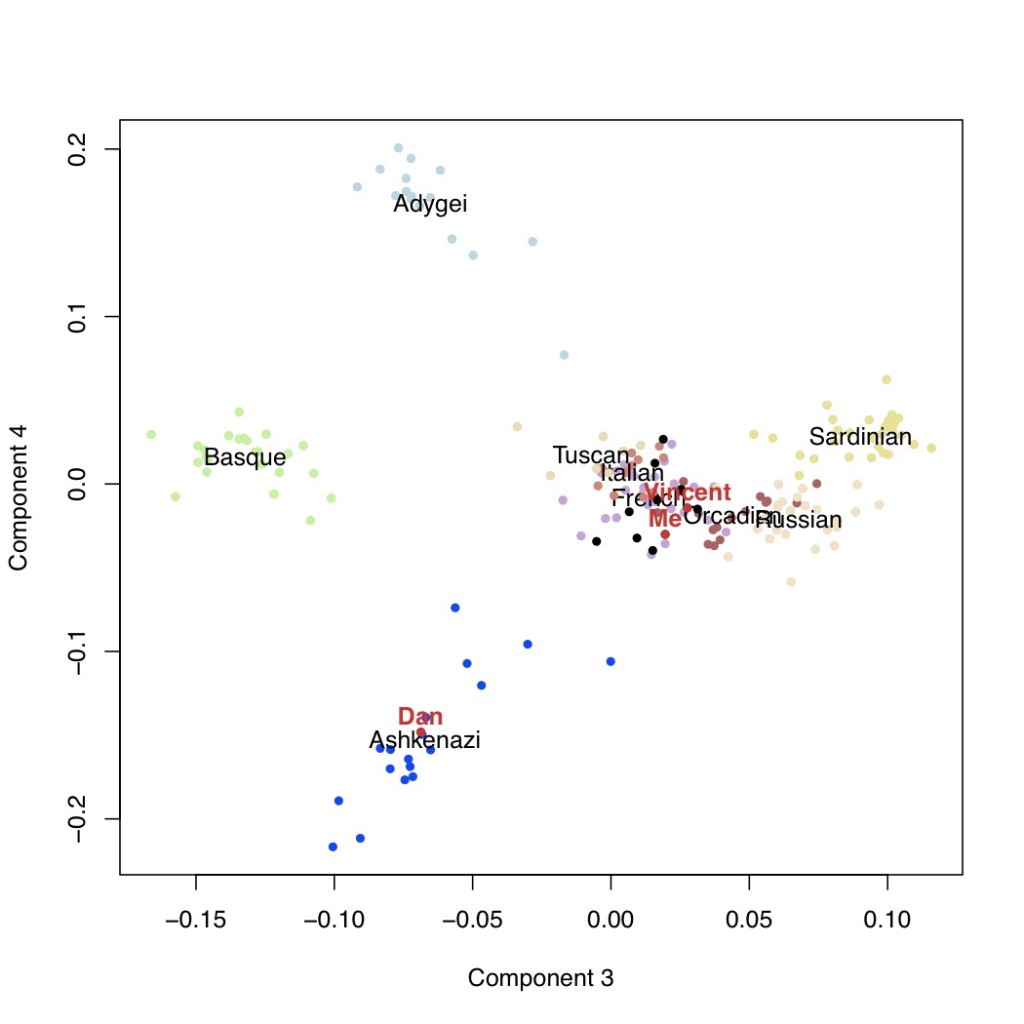
It is usually thought that we can confidently say that if our genotyping results say that we carry a certain genetic variant, that we actually do carry that variant. However, why does this not mean that we can be confident about the prediction about disease risks?
There are many risks and benefits associated with population screening, but it turns out that the results of screening tests, whether they be genetic or otherwise, might not actually provide us with a definitive diagnosis.
Usually, an individual’s test results will categorize them as being either high or low risk for getting a disease. Here, we will explain the ways in which we actually assess the predictive ability of a diagnostic test and why the results that we receive from these tests are simply estimations and not guarantees.
In order to get an idea of the validity of a screening test, it is important to look at the problem from the opposite perspective. Before you look at how probable that it might be for someone to have a particular genotype, you should look at the probability of someone developing it if they have the specific genotype.
If there were a population of 100 people, we know from epidemiological studies that the prevalence of a condition, for this case we will use Madeupitis as an example, in this population is 5%. This means that 5 of the 100 people will develop Maudeupitis, and the remaining 95 people will not.
We can summarise the screening results in the diagram pictured below, and the people colored in blue will develop the disease, and the people colored in green will not develop the disease. The red dot will indicate a positive test result.
Through this test, we can identify that 42 people are likely to develop Madeupitis, and from these people, 4 of them will be true positives, and the rest of the people will be false positives. The positive Prediction value is the number of people with a positive test result that actually go on to develop the condition.
In this case, it is 0.095 of the test population. The test will also categorize 58 people as being unlikely to develop Madeupitis, and one of these will be a false positive. The Negative Prediction value is the number of people with a negative test result that do not develop the condition, which, in this case, is 0.983.
This tells us that the test isn’t accurately identifying cases, and if you are given a positive result, there is no confidence that you will go on to develop it. Essentially, it is not an accurate prediction.
However, if you are given a negative result, you are more than likely not going to develop the condition, and this can be said with confidence. This is problematic for those that are given a positive result, which may, in fact, be false.
This does not mean that a screening test cannot be useful, but it does mean that the accuracy cannot be guaranteed with more complex conditions that have not yet been thoroughly explored. It is highly likely that the predictive accuracy of these tests will continue over time through research and additional testing and screening.