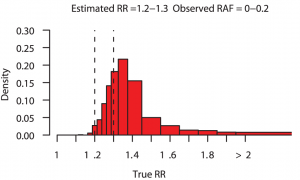
 Even though genome-wide association studies (GWAS) have identified many loci associated with complex disease, much disease heritability is still unexplained, or “missing”. But what if rather than being missing, some of the heritability was “disguised”. This is the term put forward by Chris Spencer and collegues to describe the proportion of heritability that we miss because SNPs (imperfectly) correlated to the causal variant (“tag SNPs”) are used to estimate explained heritability rather than causal variants themselves. Reassuringly, their simulations show that for the vast majority of loci detected via GWAS the risk estimated from the best tag SNP is very close to the truth. They also show that, occasionally, fine mapping of GWAS loci will identify causal variants with considerably higher risk and this is more likely if the true effect of the locus is large. The figure above, taken from their paper, shows that for estimated relative risks in the range 1.2–1.3, there is approximately a 38% chance that the true relative risk exceeds 1.4 and a 10% chance that it is over 2. The consequence of all of this for personal genomics is that disease risk could be much greater than currently thought for those individuals who, for a given disease, carry a large number of common risk variants. [CAA]
Even though genome-wide association studies (GWAS) have identified many loci associated with complex disease, much disease heritability is still unexplained, or “missing”. But what if rather than being missing, some of the heritability was “disguised”. This is the term put forward by Chris Spencer and collegues to describe the proportion of heritability that we miss because SNPs (imperfectly) correlated to the causal variant (“tag SNPs”) are used to estimate explained heritability rather than causal variants themselves. Reassuringly, their simulations show that for the vast majority of loci detected via GWAS the risk estimated from the best tag SNP is very close to the truth. They also show that, occasionally, fine mapping of GWAS loci will identify causal variants with considerably higher risk and this is more likely if the true effect of the locus is large. The figure above, taken from their paper, shows that for estimated relative risks in the range 1.2–1.3, there is approximately a 38% chance that the true relative risk exceeds 1.4 and a 10% chance that it is over 2. The consequence of all of this for personal genomics is that disease risk could be much greater than currently thought for those individuals who, for a given disease, carry a large number of common risk variants. [CAA]
Over at Genomics Law Report, Dan Vorhaus argues that direct-to-consumer (DTC) genetics companies like 23andMe will find it increasingly difficult to market the potential health relevance of their tests while simultaneously claiming that they are not providing medical advice:
When it comes to DTC genetic testing, nothing is certain. Still, we think it likely that one consequence of closer regulatory oversight will be that DTC companies are forced to choose, and to much more clearly convey to their potential customers, whether they are offering a clinical service (e.g., one designed to provide clinically useful information or to otherwise affect the individual’s health or well-being) or merely an informational service (e.g., one designed to provide access to personalized genetic information, possibly in conjunction with certain interpretive tools). The former is certain to be far more tightly regulated than the latter, at least at the outset. [DM]
This week the Wellcome Trust Sanger Institute announced the Deciphering Developmental Disorders project, a joint initiative of the Wellcome Trust and UK Department of Health. In collaboration with all of the 23 Clinical Genetics Services in the UK, the DDD project will integrate data from array-CGH, SNP arrays, and next-generation sequencing to provide diagnoses and care to patients with severe physical or mental developmental delay, or physical malformations. This work will lead to the expansion of DECIPHER, a database of variants identified in patients with developmental disorders, which will facilitate both clinical interpretation of, and scientific research into, these conditions. A core component of the DDD project is also an examination of the ethical issues facing clinicians, scientists, patients and their families as genomic technology is integrated into clinical medicine [Disclosure: Jeff, Caroline, and I all work on the DDD project; KIM].








 RSS
RSS Twitter
Twitter
0 Responses to “‘Disguised’ heritability, changes ahead to marketing of personal genomics, deciphering developmental disorders”