Though this site is largely dedicated to discussions of personal genomics, I’d like to use this post to discuss some of my recent work (done with Athma Pai, Yoav Gilad, and Jonathan Pritchard) on mRNA splicing. Our paper, in which we argue that splicing is a relatively error-prone and noisy process, has just been published in PLoS Genetics [1].
Some background
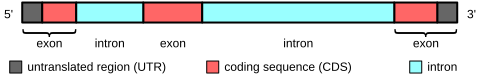
As many readers of this site know, most human genes are encoded in the genome in a bizarre fashion: the protein-coding parts of the gene are split into small chunks (called exons) separated by large swathes of non-coding, largely useless DNA (called introns; see the figure above). In order to fashion a functional mRNA (and thus a functional protein) from this type of organization, the cell first transcribes a long pre-mRNA, then decides which parts of the pre-mRNA are exons and removes the remainder via a process called splicing.
Though the process of splicing is somewhat convoluted (and was likely slightly deleterious when it initially evolved in the ancestor of all eukaryotes), it can be regulated by the cell in clever ways such that the same gene can produce different proteins in different conditions via alternative splicing. Importantly, genetic variation between individuals can also influence splicing.
Earlier this year, we published a paper in which we used high-throughput sequencing of mRNA in about 70 individuals to, among other things, try to identify the precise genetic variants influencing variation in splicing between individuals [2]. In the course of doing this, we developed methods for identifying previously unobserved splice forms. Using these methods, we saw something that was then (to us) somewhat perplexing: an abundance of never-before-seen splice junctions and splice forms in nearly every gene we examined. This paper presents the follow-up work on that observation.
What do we show?
After polishing our methods a bit more, we ultimately identified about 300,000 splice junctions in our data, about half of which had never before been observed. These splice forms are generally at low abundance in the cell and show no evidence of evolutionary conservation. Our conclusion, then, was that we are measuring the error rate of splicing reactions on a genome-wide scale.
Doing a back-of-the-envelope calculation with these data, we estimate that the error rate of the average splicing reaction in the human genome is about 0.7%. This works out to a few percent of transcripts from the average gene being mis-spliced (since most genes undergo multiple splicing reactions).
This might seem like a rather high error rate. However, consider that tens or hundreds of bases are necessary for the fully efficient removal of an intron, and that a mutation that disrupts any of these bases can cause a reduction in the efficieny of the reaction. Every generation, these mutations occur, and if they’re not sufficiently deleterious, it’s inevitable that some will reach fixation and be carried by all humans. This idea is not new, of course; Michael Lynch has referred to this fact as part of the “intrinsic cost of introns” [3]. But what we’ve shown is that these new sequencing technologies allow us to measure these sorts of things on a much larger scale than was previously possible.
—-
[1] Pickrell et al. (2010) Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genetics.
[2] Pickrell et al. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. doi:10.1038/nature08872.
[3] Lynch (2007) The origins of genome architecture. Sunderland, Mass.: Sinauer Associates.








 RSS
RSS Twitter
Twitter
This is interesting – it makes some sense as well but who would have predicted it?! DNA replication and repair are themselves fairly error prone but there is a complex, expensive?, setup to proof-read and correct errors to keep them to the minimum required for life leaving a bit of mutation useful for evolution. I suppose that errors in splicing, as long as they are kept to a low level, as your work suggests, are not so dangerous.
Very interesting paper. I am curious to hear your take on the importance of RT polymerase errors in this type of experiment. Couldn’t RT slippage make cDNAs that would look spliced? At what rate would this happen? I suppose most cases would not have GT-AG and so not be in your study but I am wondering how many of these you found (thats the 1.5%)?
congrats on the paper mr. pickrell.
a little off topic, has anyone ever seen a PLoS paper where the comments ever got hoppin’?
txs. never seen a paper with a lot of comments, but I’ve seen a paper with a damning comment; this sort of things speaks to the utility of a commenting system:
http://www.plosone.org/annotation/listThread.action?inReplyTo=info:doi/10.1371/annotation/5f65ab1c-9c77-4376-b1e5-88053db2dff4&root=info:doi/10.1371/annotation/5f65ab1c-9c77-4376-b1e5-88053db2dff4
greg, thanks for the comment. the short answer is I’m not sure how important RT polymerase slippage is. presumably that sort of thing would lead to relatively short “introns”? ie. it’s unlikely that the polymerase would slip 10kb; is that right? One thing I guess we didn’t mention in the paper is that we removed spliced reads that lead to very short “introns”, which we assumed were due to either sequencing or mapping errors.
Thats what I also thought when I first started looking at this. I was very surprised by the recent work by McManus (et al 2010), where they found lots of evidence for template switching/ RT errors. Looking at your study though it does not seem to be as frequent as I was thinking, as most RT artefacts should be in that 1.5% with out GTAG. Its just something I do not see mentioned very often in the literature and I am trying to figure why that is.
McManus CJ, Duff MO, Eipper-Mains J, Graveley BR. Global analysis of trans-splicing in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20615941.
greg, thanks for that link. We limited ourselves to spliced reads were both ends mapped 1) on the same strand and 2) within 20kb of each other, which I think helped us a lot. We did get quite a few reads where the ends mapped to different chromosomes or very far apart. My instinct was that these were errors in mapping, but it’s also possible they were polymerase errors.