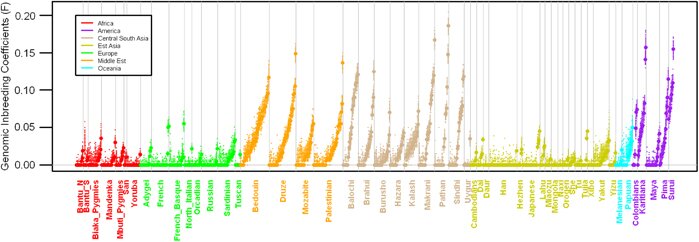
 The picture above shows the relatedness of parents across different populations throughout the world; for reference, 0.065 is the average value for a first cousin mating. It comes from a paper out in pre-publication this week at the European Journal of Human Genetics, which estimates the level of inbreeding (or “consanguinity”) of parents by looking for sections of the genome where individuals inherit an identical piece of DNA from each parent. Such “runs of homozygosity” are a sure sign of inbreeding, as both parents will have inherited the bit of DNA from a recent common ancestor: the number and length of these sections can be used to find out how many generations ago the common ancestor lived, i.e. how closely related the parents were (cousins share a grandparent, second cousins a great-grandparent, and so on). In the plot above, we can see a high degree of cousin-marriage in Middle-Eastern cultures, and somewhat more sadly, high degrees of inbreeding in the Native American populations, due to the collapse in their population sizes. [LJ]
The picture above shows the relatedness of parents across different populations throughout the world; for reference, 0.065 is the average value for a first cousin mating. It comes from a paper out in pre-publication this week at the European Journal of Human Genetics, which estimates the level of inbreeding (or “consanguinity”) of parents by looking for sections of the genome where individuals inherit an identical piece of DNA from each parent. Such “runs of homozygosity” are a sure sign of inbreeding, as both parents will have inherited the bit of DNA from a recent common ancestor: the number and length of these sections can be used to find out how many generations ago the common ancestor lived, i.e. how closely related the parents were (cousins share a grandparent, second cousins a great-grandparent, and so on). In the plot above, we can see a high degree of cousin-marriage in Middle-Eastern cultures, and somewhat more sadly, high degrees of inbreeding in the Native American populations, due to the collapse in their population sizes. [LJ]
For those interested in gene-by-environment interactions the latest issue of Trends in Genetics includes a review article by Carole Ober and Donata Vercelli on the challenges of this area, illustrated by examples from asthma research. In particular they highlight the difficulties of moving G-by-E studies from examination of known candidate genes to genome-wide association. More interactions, this time of the protein-by-protein kind, are the subject of an article by Soler-Lopez et al. in this month’s Genome Research. They looked for interactions relating to Alzheimer disease using a combination of computational and experimental strategies, identifying 66 genes that putatively interact with known AD-related genes. The authors focus on the potential roles of neuronal death regulation and pathways linking redox signalling to immune response in AD pathology. [KIM]
This week, Debbie Kennett pointed out two separate indicators of the growing numbers of people beginning to engage with their genetic information. Firstly, an article in the Mercury News notes that an astonishing 470,000 people have now participated in The Genographic Project, which provides information on a relatively small number of markers on the Y chromosome and mitochondrial DNA to infer an individual’s deep ancestry. Secondly, a more modest but nonetheless impressive figure: 23andMe’s Alex Khomenko noted on Quora that more than 75,000 individuals are in 23andMe’s database as of last month. [DM]
Charles Warden has posted a largely positive three-part review of his experience undergoing testing by 23andMe. He first compares his 23andMe interpretation with the results provided by the free software Promethease; then emphasises the need to consider genetic results in the context of other risk factors; and finally, he takes issue with 23andMe’s presentation of trait prediction. While Warden’s point about the need to consider environmental as well as genetic risk factors is important, it’s worth noting that there currently isn’t enough information about the interactions between genes and environment to build all of these factors into a single predictive model (as noted in the article by Carole Ober discussed above by Kate). [DM]
 Genomes Unzipped content is available for reuse under a Creative Commons Attribution-ShareAlike 3.0 Unported License.
Genomes Unzipped content is available for reuse under a Creative Commons Attribution-ShareAlike 3.0 Unported License.








 RSS
RSS Twitter
Twitter
Yes, the Ober-Vercelli review is excellent. The challenges facing the GWAS arena in identifying GxE interactions are significant. To try to predict where GxEs are likely to be found, we start with those that have been determined experimentally. That is facilitated with a catalog of known GxEs such as those we’ve put together in a paper just released at http://omicsonline.org/2153-0602/2153-0602-2-106.pdf.
Hi,
I don’t want to be overly critical, and maybe I’m mis-understanding the
target audience for these posts, but I have to say that hitting a phrase
such as ‘..which estimates consanguinity of parents using runs of homozygosity
in their children.’ in the second sentence of the article was..jarring to say
the least. I *think* I know what you mean but surely there was a way of
writing it in plain english ?
Ed Yong (among others) has written extensively on the problem of trying to
convey highly technical information to a lay or non expert audience without
large amounts of jargon:
http://blogs.discovermagazine.com/notrocketscience/2010/11/24/on-jargon-and-why-it-matters-in-science-writing/
Just a thought.
Yes, a very good point. I wrote that very rapidly, and was pretty lazy in my use of language. I have updated it to explain in more detail what the paper is looking for.
Thanks for pointing that out.
Bonjour,
Thanks for citing our paper.
The method estimates the level of inbreeding of the individuals or equivalently the level of kinship between their parents, but not the level of inbreeding of parents.
Maybe something along those lines would read better “which estimates the level of inbreeding (or “consanguinity”) of individuals by looking for sections of the genome where the two parental chromosomes have identical DNA”.
@Anne-Louise
Thanks for pointing that out! I’ve modified it to be a bit less wrong.
This post was timely. I just calculated my % heterozygosity of 23 and me markers. I find I’ve got about 90% of the variation of the genomes unzipped authors, who were all more heterozygous than me. Perhaps it’s my notoriously consanguineous ethnicity of origin (which I see is somewhat less sad than the decimating population collapse in Native American populations) or perhaps it’s more an issue of cosmopolitan vs rural effects on mate choice. I’d love to see a paper on the strength of the effect of cosmopolitanrural living on calculations of individuals’ genetic variation.
@Mary
Worth remembering that there is (sort of) a difference between lack of homozygosity and runs of homozygosity. Dan is far more homozygous than the rest of us, but that is a result of his Ashkenazi ancestry, not any recent relatedness of his parents.
Here’s the raw data:
file: CAA001_genotypes.txt heterozygosity: 30.8358275538311
file: CFW001_genotypes.txt heterozygosity: 31.4044454411371
file: DBV001_genotypes.txt heterozygosity: 30.7424563878846
file: DFC001_genotypes.txt heterozygosity: 30.80868075188
file: DGM001_genotypes.txt heterozygosity: 30.5667802682519
file: IPF001_genotypes.txt heterozygosity: 31.4325384680558
file: JCB001_genotypes.txt heterozygosity: 30.8904669768664
file: JKP001_genotypes.txt heterozygosity: 31.0774311799696
file: JXA001_genotypes.txt heterozygosity: 30.7756550246656
file: KIM001_genotypes.txt heterozygosity: 31.398475283442
file: LXJ001_genotypes.txt heterozygosity: 30.542745838499
file: VXP001_genotypes.txt heterozygosity: 30.7749633863993
file: genome_Mary_Paniscus_Full_20110201104429.txt heterozygosity: 29.5017358220516
and the script I used to get it:
**********************************************
use warnings; use strict;
unless($ARGV[0]){die “usage: perl $0 *.txt\n”}
my @files=@ARGV;
foreach my $file (@files){
chomp $file;
open (FH, $file);
my $het = 0;
my $total = 0;
while (my $line = ){
if ($line=~/^#/){next}
$total++;
chomp $line;
my @elements=split(/\s+/,$line);
my ($first,$second)=split(”,$elements[3]);
unless ($first=~/[AGTC]/){next}
unless ($second=~/[AGTC]/){next}
unless ($first){next}
unless ($second){next}
if ($first eq $second){next}
$het++;
}
my $percent=$het*100/$total;
print “file: $file heterozygosity: $percent\n”;
}
**********************************************
It just calculates the % heterozygous markers for each file. I haven’t changed it to only use markers shared by all the files under consideration yet. So these values are obviously affected by using a different gene chips.
Hi Mary,
Is your data from the v3 chip? If so, it’s very likely that this explains at least some of the difference: we’re all on v2, and the extra SNPs on the v3 are probably enriched for rare SNPs that will show lower heterozygosity.
I like this result better ;) –though I’m not finding Dan has the lowest heterozygosity.
of 520288 total shared markers
file: CAA001_genotypes.txt heterozygosity: 31.8610077495541
file: CFW001_genotypes.txt heterozygosity: 31.8291023433175
file: DBV001_genotypes.txt heterozygosity: 31.7943139184452 <- Dan V
file: DFC001_genotypes.txt heterozygosity: 31.8287179408328
file: DGM001_genotypes.txt heterozygosity: 31.6426671382004 <- Dan M
file: IPF001_genotypes.txt heterozygosity: 31.7768236053878
file: JCB001_genotypes.txt heterozygosity: 31.951918937204
file: JKP001_genotypes.txt heterozygosity: 32.188710867827
file: JXA001_genotypes.txt heterozygosity: 31.8477458638293
file: KIM001_genotypes.txt heterozygosity: 31.7710575681161
file: LXJ001_genotypes.txt heterozygosity: 31.6222938065072 screened”);
my $line_count=0;
foreach my $file (@files){ #1st step: screen lines
chomp $file; # print “file: $file\n”
open (FH, “$file”);
while (my $line = ){
if ($line=~/^#/){next} #no comment lines
chomp $line;
my @elements=split(/\s+/,$line);
if ($#elements screened.sorted”);
#initialize counters
my $total = 0;
my %het; foreach my $f (@files){$het{$f}=0}
open (FH2,”screened.sorted”);
#2nd step: count het sites of only shared sites (use same id#)
my $lines_counted=1;
my $last_line=;
my @lines = ($last_line); #where we put all the lines matching last line’s id
while ($lines_counted < $line_count){
my $next_line=; $lines_counted++;
#print “last: $last_line”, “next: $next_line”;
my ($id,$chr,$pos,$nt,$file_name)=split(/\s+/,$last_line);
my ($id2,$chr2,$pos2,$nt2,$file_name2)=split(/\s+/,$next_line);
#print “id:*$id* id2:*$id2*\n”;
#foreach my $line(@lines){print “in lines array:$line\n”}
if ($id eq $id2){push(@lines,$next_line)}#add to list of same ids
else { #if not a match
$last_line=$next_line; #put in last line for next round
#print “#lines:$#lines #files:$#files\n”;
unless ($#lines eq $#files){@lines = ($last_line);next} #do we have reps for all genomes?
$total++;
foreach my $line(@lines){
my ($id,$chr,$pos,$nt,$file_name)=split(/\s+/,$line);
my ($first,$second)=split(”,$nt);
if ($first eq $second){next}
$het{$file_name}++;
}
@lines = ($last_line);
}
}
print “of $total total shared markers\n”;
foreach my $file (@files){
my $percent=$het{$file}*100/$total;
print “file: $file heterozygosity: $percent\n”;
}
ouch, that really did get maimed up there: here are the complete results with no attempted script attachment:
of 520288 total shared markers
file: CAA001_genotypes.txt heterozygosity: 31.8610077495541
file: CFW001_genotypes.txt heterozygosity: 31.8291023433175
file: DBV001_genotypes.txt heterozygosity: 31.7943139184452 <- Dan V
file: DFC001_genotypes.txt heterozygosity: 31.8287179408328
file: DGM001_genotypes.txt heterozygosity: 31.6426671382004 <- Dan M
file: IPF001_genotypes.txt heterozygosity: 31.7768236053878
file: JCB001_genotypes.txt heterozygosity: 31.951918937204
file: JKP001_genotypes.txt heterozygosity: 32.188710867827
file: JXA001_genotypes.txt heterozygosity: 31.8477458638293
file: KIM001_genotypes.txt heterozygosity: 31.7710575681161
file: LXJ001_genotypes.txt heterozygosity: 31.6222938065072 <—–lowest, also Jewish?
file: VXP001_genotypes.txt heterozygosity: 31.7914308998093
file: genome_Mary_Paniscus_Full_20110201104429.txt heterozygosity: 32.1912094839781
I rewrote my earlier script to only use the subset of markers shared by all individuals files, and to exclude those on X and Y chromosomes. I sent it to the blog's gmail.
Thanks for your post on this blog site. From my own experience, many times softening way up a photograph could possibly provide the digital photographer with a bit of an artistic flare. Often however, the soft cloud isn’t precisely what you had under consideration and can often times spoil an otherwise good picture, especially if you thinking about enlarging that.