This is the first of a new format on Genomes Unzipped: as we acquire tests from more companies, or get data from others who have been tested, we’ll post reviews of those tests here. The aim of this series is to help potential genetic testing customers to make an informed decision about the products on the market. We’re still tweaking the format, so if you have any suggestions regarding additional analyses or areas that should be covered in more detail, let us know in the comments.
Overview
Lumigenix is a relative newcomer to the personal genomics scene: the Australian-based company launched back in March this year, offering a SNP chip-based genotyping service similar in concept to those provided by 23andMe, deCODEme and Navigenics.
The company kindly provided Genomes Unzipped with 12 free “Comprehensive” kits, which provide genotypes at over 700,000 positions in the genome, to enable us to review their product. We note that the company offers several other services, including a lower-priced “Introductory” test that covers fewer SNPs, and whole-genome sequencing for the more ambitious personal genomics enthusiast. This review should be regarded as entirely specific to the Comprehensive test.
Technology
The Lumigenix Comprehensive platform is based on Illumina’s HumanOmniExpress+, the same chip used by 23andMe. Lumigenix test for around 732,000 standard variants, along with around 2,700 specially selected ones, which appear to be mostly mitchondrial or Y chromosome markers. This is somewhat less than 23andMe (734,000 variants plus around 230,000 custom ones) and deCODEme (1.1 million variants).
Pre-purchase information
Lumigenix do try to make the limitations of genetic risk projections clear in their pre-purchase information and explicitly differentiate themselves from clinical genetic testing services. They focus on the advantages of awareness and information as a step to enabling individuals to improve their own health, if they choose, and make it clear that genetic testing alone does not accomplish this. White papers describe their approach to health risk calculation (database curation and algorithm – PDFs) as well as ancestry calculations (PDF).
Information regarding the limitations of the service offered is detailed in one of the information pages, but only there is only a scant discussion of how many variants are included for different ethnic groups. Although it is possible to select from 10 different ethnicities, it is hard to find out exactly what the ramifications of this are without going to the page of each trait individually. It would be much more transparent if all DTC companies followed Pathway Genomic’s approach and provided a list of conditions with ethnicities covered. Ideally, information would also be available about the predictive capacity of the variants included for each ethnicity as well.
The testing experience
The Lumigenix testing experience will be familiar to anyone who has tested with virtually any of the major personal genomics companies: the testing kit that arrives in the mail is basically the same boxed Oragene saliva collection kit that 23andMe provides, albeit somewhat more plainly packaged. The spit kit itself is straightforward to use: spit in the tube until you reach the line, then close the hinged lid to dump preservative into the tube, and then finally replace that lid with a permanent screw-on lid for return to the company.
One small difference between the Lumigenix and 23andMe kits is that Lumigenix tucks the return envelope and biohazard bag in a separate compartment in the kit box – several of us missed this entirely and had to be guided by other (more perceptive) members.
Error rates
We’ve taken a number of different approaches to estimate error rates for the various platforms that have been used by the group so far, only one of which is presented here (Luke will have more details in an upcoming post). For this analysis we took advantage of the fact that one individual in the group (Caroline) has been genotyped independently on three separate platforms (23andMe, deCODEme and now Lumigenix), and that there are around 329,000 sites that are assessed by all three companies. In this three-way comparison we looked for sites where two companies agreed on Caroline’s genotype and the third disagreed; we assume that such sites represent genotyping errors by the third company.
There is a major caveat to this analysis: sites that are shared between all three platforms are not necessarily a representative sample of the sites on any one of the chips – in fact, it’s extremely likely that these sites will be biased towards more common and better-behaved markers (i.e. this analysis likely underestimates the true error rate). Consistent with this, a less biased approach that Luke has performed on the 23andMe platform (using data from himself and his parents) suggests that the overall error rate for the 23andMe chip is substantially higher than the number above suggests, perhaps closer to 13 errors per 100,000 sites. As we don’t have family data for Lumigenix we can’t perform this analysis here.
Anyway, given that caveat, Caroline’s different genotype data-sets do tell us two interesting things about the calling approaches taken by the three companies, at least for these 329,000 shared sites. Firstly, Lumigenix are tend to be more zealous in their genotype calling, giving non-missing genotypes for 99.8% of variants (compared to 99.3% and 98.5% for 23andMe and deCODEme respectively) – in other words, if there is uncertainty at a site they are more likely to provide a “best guess” genotype rather than assign the site as “unknown”. Secondly, while error rates for all chips are extremely low, Lumigenix has a higher error rate than either of the other two: 7.4 errors per 100,000 variants, compared to 2.5 and 1.9 for 23andMe and deCODEme respectively. This is almost certainly because Lumigenix is more likely to make an educated guess at hard-to-genotype sites, so while it makes more guesses, it also gets more of thme wrong.
(We’re currently in the process of collecting the group’s raw data from the Lumigenix test, which will be posted here.)
Health
Summary
Results are returned for 81 heath-related conditions. Lumigenix essentially use a pared-down version of the now-familiar 23andMe-style of risk communication. Traits are split into high, low, and average risk for an individual and for each condition your risk, the average risk and your relative risk, and the strength of evidence behind the association, are reported. The strength of evidence is a five point scale where 1-4 points indicates a preliminary finding and 5 points indicates an ‘established’ finding, defined as follows:
The highest evidence rating is given to established condition reports, where at least two independent studies have demonstrated an association between the same sets of SNPs. A lower evidence rating is assigned to associations built from single studies.
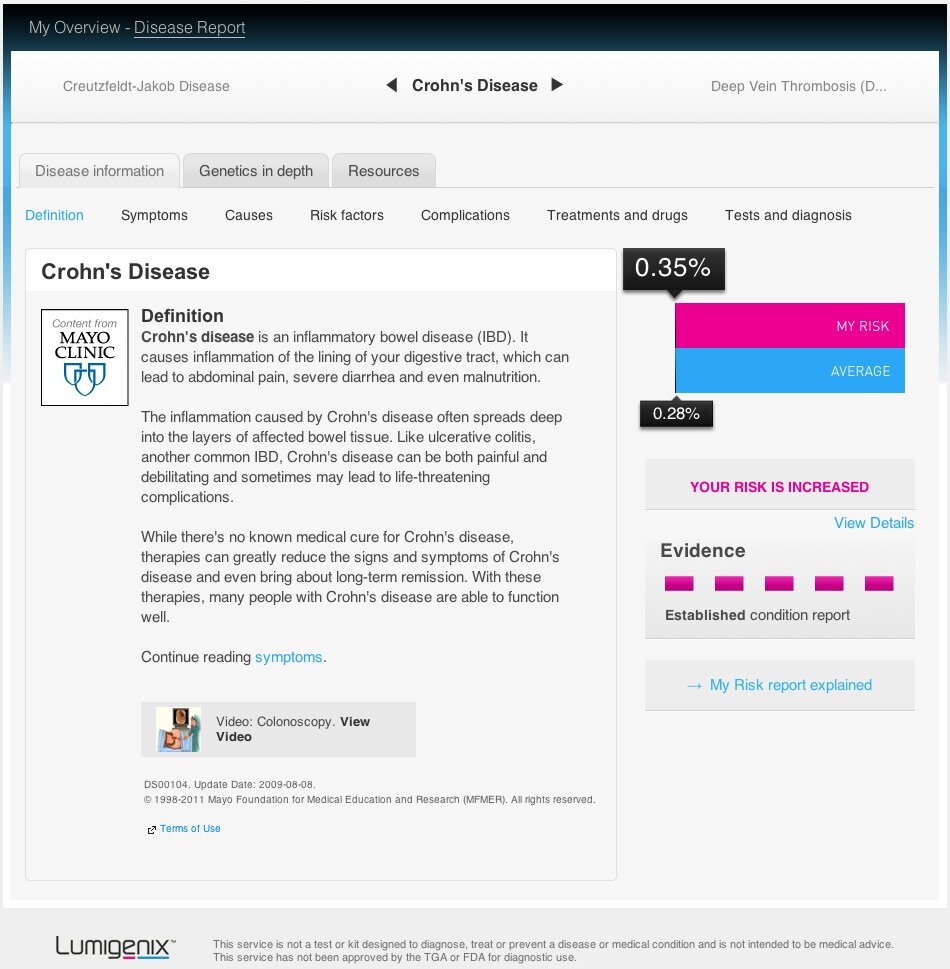
The user can than click through to a disease-specific page (see example below) containing information from the Mayo Clinic on disease symptoms, causes, risk factors, complications, treatments, and clinical tests for diagnosis. From this page it is also possible to retrieve the information on which SNPs were tested and the user’s genotype and odds ratio, plus references for all the SNPs included.
On the positive side, Lumigenix’s collaboration with the Mayo Clinic means that they are able to make quite a bit of medical information available about each “Heath” association. Additionally, for those who are interested in the information going into Lumigenix’s risk prediction, the details of the SNPs included are easily accessible from the “Genetics In Depth” tab for each “Health” association. However, the “Health” pages lack many of the various visual guides to interpretation that 23andMe use both to convey information and make the experience more entertaining.
Evidence supporting disease association
Most of you who have followed the personal genomics field will be aware of the fact that predicted risks can differ substantially between companies, for a number of reasons – some of which are mundane and depressingly preventable, while others are more difficult to decipher.
Comparing our Lumigenix and 23andMe results provided plenty of examples of such discrepancies: for example, Joe’s 23andMe results give him a much higher risk for prostate cancer than Lumigenix. Why the difference? Well, it’s hard to say for sure. 23andMe has more markers typed for this disease, despite Lumigenix citing the same papers. When you click on a variant in 23andMe, it tells you the precise references for that individual variant, whereas Lumigenix only gives a list of general references for the disease. That makes it very difficult to figure out exactly which references have been used to define the frequencies and effect sizes for each variant and identify the source of the discrepancies.
We believe it’s essential that personal genomics companies make the basis for their risk calculations as transparent as possible to allow customers to fully explore the evidence supporting a company’s tests. While Lumigenix does better than some companies in the industry by actually providing references at all, it could do better: we would like to see explicit links between each variant and the corresponding reference.
Completeness of tested markers
Lumigenix claim to have better coverage of common diseases, compared to 23andMe and deCODEme, based on number of SNPs genotyped. However, it’s important to note that it’s not just the number of SNPs genotyped that matters, but also which SNPs are selected. One way we can evaluate this is to use a number called the AUC (for “area under the curve”) for a set of variants. The AUC is a number between 0.5 and 1 that measures how well a test classifies individuals with and without a disease: a value of 0.5 suggests that the test has no ability to predict disease (i.e. you might as well flip a coin), and the cluser the value is to 1 the better the test is (for more information see Wray et al. 2010).
As an example, we examined the SNPs included for Crohn’s disease (CD), a well-studied common complex disease, from Lumigenix, 23andMe, and deCODEme. Luke identified all published markers with a replicated genome-wide significant association with CD (note: we did not require that the replication study be completely independent), and calculated the variance explained using the sets of variants each company uses. In each case, he used the odds ratios and frequencies from the Franke et al. (2010) meta-analysis to calculate variance explained by this variant set:
| Variant set | Number of variants | Variance explained | AUC |
| Luke’s “best set”: variant with lowest p-value in each CD region plus NOD2 variants | 73 | 11.4% | 0.76 |
| Lumigenix set (includes 1 NOD2 variant) | 16 | 5.1% | 0.68 |
| 23andMe set (includes 3 NOD2 variants) | 12 | 5.5% | 0.69 |
| deCODEme set (no NOD2 variants) | 30 | 6.0% | 0.70 |
The main thing to take away from these data is that all three sets of SNPs from the DTC companies have about the same AUC, despite including different numbers of SNPs. 23andMe has a carefully hand-curated database of risk variants, and have designed their chip to optimally tag these. They have specially included the three major mutations in the important gene NOD2, which make up about 20% of the known genetic risk for Crohn’s disease. However, their set is also much smaller, so only 12/73 known variants are included. At the other extreme, deCODEme has not included any of the NOD2 mutations, but has included many more of the other variants. Lumigenix is somewhere between the two, including one of the NOD2 mutations, and a few more of the other variants. Notice that none of the companies actually do as well as they could here, as none of them include both the NOD2 mutations, and all the other variants (though 23andMe could, if they updated their database accordingly).
Traits
The user also receives results for nine “traits”: alcohol flush reaction, bitter taste perception, caffeine metabolism, earwax type, episodic memory, eye colour, freckling, fullscale IQ, and hair colour. These are simply reported as the likely outcome (e.g. “Likely brown or black hair”), and clicking through to the trait page brings up a bit of information about the trait and the specific SNPs tested. It’s not clear whether the associations reported here are subject to the same strength-of-evidence standards as the health-related results.
Ancestry
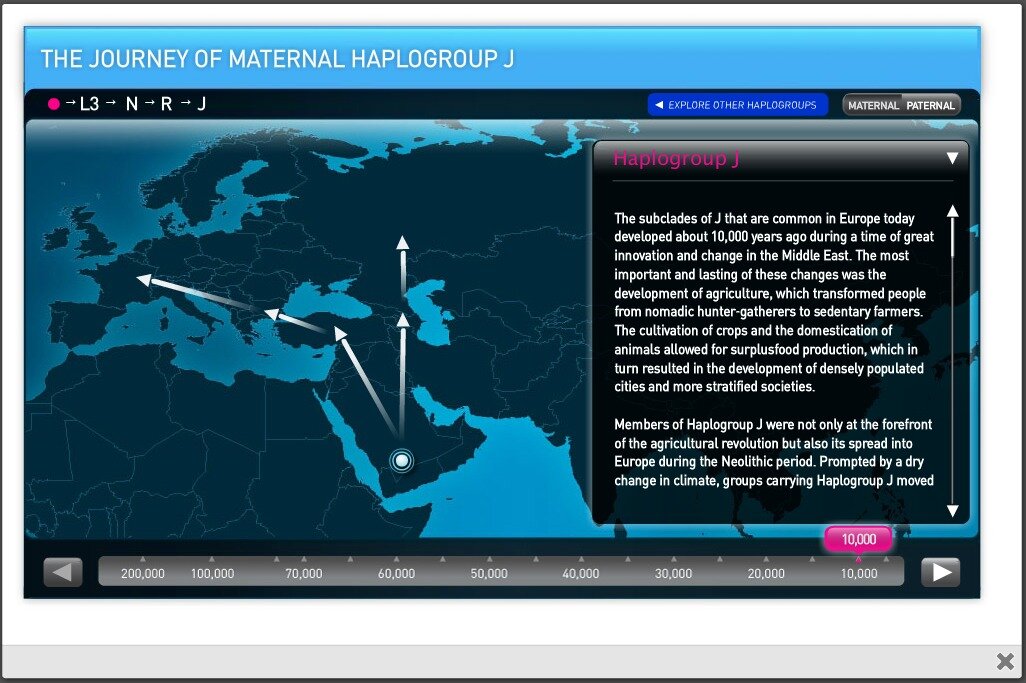
Lumigenix have put some effort into making their ancestry section both informative and entertaining. Their human migration map (see screenshot below) traces the historical movement of different haplogroups in an interactive way.
However, while the map is pretty, it’s also curiously disconnected from the rest of the site – rather than automatically zooming in to show your specific haplogroups, you must first browse through your ancestry information, remember your haplogroups (e.g. I2 and R1a1a), then click on the map and manually find your own haplogroup from continent-specific lists. This could certainly be made more user-friendly.
Lumigenix also provide chromosomal ancestry mapping – diagrams indicating which chunks of your genome come from which continent – which seems to work reasonably well. And then, rather surprisingly, that’s where the ancestry information ends. More detailed analyses of autosomal markers (such as a principal components analysis-based map) are not available, and as sharing between users is not currently supported, there is no facility for sharing data with relatives, or identifying new ones in the user population.
Data portability
We generally believe that it is the right of personal genomics customers to have complete access to any raw genetic data generated by a company, and the responsibility of companies to provide such data in a usable form. Lumigenix does well on this front: they provide customers with the ability to freely download their complete genotype data for third-party analysis, and the company was also responsive to our suggestions regarding changes to their raw data format. The current raw data format (as of 30th November 2011) is a tab-delimited file containing fields for marker ID, chromosome number and position (relative to the build 36 reference sequence), customer genotype, and a final column indicating the two possible alleles for that marker.
Further improvements to the data format could certainly be made: for instance, a header specifying the coordinate system used, the date the file was downloaded and the field titles would be useful, and it would probably minimise confusion if the right-most column contained the customer genotype rather than the two possible alleles. However, in the absence of any industry-wide consensus on raw data format it’s hard to be too picky, and we think Lumigenix has done a comparable or better job than most of the other companies in the market in terms of providing usable data.
Unlike 23andMe, but similar to pretty much all other health-related personal genomics companies, Lumigenix provides no direct mechanism for sharing your genetic results with other users of the service. Buying a Lumigenix test will not allow you to find new relatives or socialise with people who share your genetically determined earwax consistency.
Overall conclusions
Firstly, we should be clear that there are many things that Lumigenix has done well, especially compared to companies occupying the lower end of the direct-to-consumer genetics market – this is absolutely not a cowboy operation. We particularly liked Lumigenix’s collaboration with the Mayo Clinic, which is a sensible way of obtaining reliable disease information without needing to invest heavily in in-house curation.
However, Lumigenix can’t help but suffer from comparisons to 23andMe, a company that beat it to market by over three years and now thoroughly dominates the direct-to-consumer genomics industry. The overall experience feels more-or-less like a less satisfying version of 23andMe: the common disease, trait and ancestry prediction systems are all broadly similar, but with a less sophisticated interface, and many of the more interesting components of the 23andMe service (such as carrier testing, pharmacogenomic markers, genome sharing, family inheritance patterns, Relative Finder) feel quite conspicuous in their absence. Then there’s the price: the Lumigenix test currently retails for USD$479 while 23andMe is USD$399 (without subscription).
We’re all for competition in the personal genomics market: new companies in the space should bring new ideas and help drive prices down. But when the new companies offer services that are basically less useful versions of those offered by the market leader, at a higher price, it’s hard not to be disappointed.
Still, nothing stands still in personal genomics – and Lumigenix has assembled a reasonably solid base from which to branch out into more innovative directions. We hope it chooses to take paths less travelled in its future products.









 RSS
RSS Twitter
Twitter
This is such an incredibly valuable service to the personal genomics community. Thank you on behalf of me and all my #PM101 students, as well as dozens of non-scientists for whom I’m attempting to convince the value of personal genomics!
Re: “if you have any suggestions regarding additional analyses or areas that should be covered in more detail, let us know in the comments”
It seems that many of the original DTC genetic testing companies are no longer DTC, thanks to the recent threats from the FDA. What might be useful is a running list of companies that remain open to the consumer (no physician required) so that consumers could rapidly compare and contrast by merely clicking among a list of hyperlinked websites. Because of flux in this rapidly moving area, something like this would facilitate ones’ “current options”.
Thanks again.
-Bob
Bob –
The most current such list I know of was pulled together by GPPC last summer: http://www.dnapolicy.org/resources/DTCTableAug2011Alphabydisease.pdf
It is tough to track all of these because the business models / product offerings do change rapidly but, in general, yes I think there was some flight from “true DTC” following the Congressional hearing, GAO report and FDA letters.
Things have been relatively quiet recently, however, so perhaps that will change in 2012. It’s hard to see the appeal for new entrants in challenging 23andMe, but perhaps DTC-focused exomic or whole-genome sequencing?
– Dan
Thanks for your rapid and precise reply, Dan!
– Bob
The post text indicates that the lumigenix data for all 12 is available, but https://genomesunzipped.org/data has not yet been updated.
Using the data from Daniel MacArthur which is online I freshened up the Promethease reports at
http://www.snpedia.com/index.php/User:Daniel_MacArthur
In particular, the ui2 reports allow a much more powerful exploration of the data
http://files.snpedia.com/reports/GNZ/promethease_data/promethease_DGM001_Daniel_MacArthur_pooled_ui2.html
Sorry Mike – I’ve deleted that statement in the post. I’ve now collected data from most of the group, and will put those files live some time next week.
Your columns are extremely interesting and generally do an excellent job of explaining technically complex information to the public. I do have some suggestions, however, that will make your writing easier to read for most people.
Please consistently use a company name – Lumigenix, for example – as a SINGULAR noun or as an adjective. As a single company, Lumigenix is after all a SINGLE entity, not a plural collection of entities. So a phrase like “Lumigenix also provide chromosomal ancestry mapping” would be more correctly written as “Lumigenix also provides chromosomal ancestry mapping”. This may seem like a small thing, but I found that your unconventional treatment of “Lumigenix” as a plural noun made the writing in this column seem forced, to the point of tortured, and almost unreadable: NOT good qualities when the reader needs to be able to focus all of his or her attention on the technical aspects of the column.
Another alternative would be to use Lumigenix as an adjective, for example: “Lumigenix services also provide … “.
In addition, while it is technically true that “data” is plural, most readers are used to considering it in its collective sense, as a singular group. So instead of “Caroline’s data do tell us two interesting things”, which is correct but a bit awkward, it might be better to say “Caroline’s data sets do tell us two interesting things” – which is easier on the reader and even more accurate in the context in which that phrase occurs.
Finally, please do not get confused about which noun corresponds to the verb. In the following phrase, the occurrence of the word ‘data’ apparently led you astray, so that you selected ‘are’ as the verb. “The main thing to take away from these data are …” But ‘data’ is NOT the subject of the sentence! The noun/verb correspondence is actually THING/IS, thus: “The main THING [subject, singular] to take away from these data IS [verb, singular] …”
I know this may seem like unimportant window-dressing to you, but when the writing is so distracting from an important and interesting message, it’s worth it to take the time to improve the presentation of the content.
Thank you!
Hi editorial commentator,
Thanks for your unusually thorough review of the language of our post. I’ve altered the “Caroline’s data do tell us” statement, as while technically correct I agree the use of data as plural is confusing to many people. I’ve also fixed the incorrect subject plurality example.
I’m less convinced of the dangers of using Lumigenix as a plural noun. While technically incorrect, I consider this a kind of poetic license – we’re emphasising that the company’s products were put together by multiple individuals, not some imaginary monolithic entity. I certainly disagree that this usage makes the text tortured, let alone unreadable – but if other readers also find this distracting, please let me know.
Thank you for a great post. I find very interesting that this company relies on build 36/hg18 of the human genome for their annotations. Build 37 has been out for three years and has many improvements to the genome. This might be understandable for an old company that wants to remain compatible with legacy data, but Lumigenix probably did not even even exist when Build 37 was made.
Companies being referred to as singular or plural is a UK vs US English issue; with the majority of the GU writers hailing from the UK or working there, (including Dan M in this) then plural seems appropriate.
Thanks for posting this review.
Have you considered sending two kits from the same person and checking the results for consistency?
Although UK usage may sometimes treat company names as plural nouns, the UK style guides are not always in agreement with that practice. If you care to look at the style guide from The Economist – certainly a persuasive authority on the subject of how to treat company names in UK English – it states: “A government, a party, a company (whether Tesco or Marks and Spencer) and a partnership (Skidmore, Owings & Merrill) are all it and take a singular verb.” (A pdf of the source is available here: http://www.frzee.com/Education/The%20Economist%20Style%20Guide.pdf)
Despite that guidance from the Economist, there is obviously some variation in how this usage is perceived by UK as compared to US readers. If the author’s intention is to emphasize a number of individuals involved in the company, why not clearly say that as ‘Lumigenix researchers’, ‘Lumigenix executives’, or whatever group it is that you wish to refer to? That would much more surely lead the reader to an understanding of the author’s intentions than a coy (and potentially confusing) allusion to the company as a plural noun. There’s nothing to be gained in this genre from a ‘nudge, nudge, wink, wink’: instead, Just Say It.
Tom,
It’s a fair question – our comparison between different genotyping platforms for the same individual gives us a reasonable idea of consistency, but in future we may well consider sending duplicate samples in for review (if sufficient kits are available).
editorial commentator,
I re-read the post with an open mind, and my honest impression is that the vast majority of readers will never even notice the singular/plural issue. I’m a guy who generally appreciates pedantry, but here I think the discussion has outlived its usefulness – so no more comments on semantics, please.
Is a fastidious adherence to asphyxiating grammatical regulations genetic based? If so, a candidate gene approach would probably start investigation at SNPs in the SLC6A4 gene, such as the 5-HTTLPR variant… http://omim.org/entry/182138#0001