[By Gokul Ramaswami and Robert Piskol. Gokul Ramaswami is a graduate student and Robert Piskol is a postdoctoral fellow in the Department of Genetics at Stanford University. Both study RNA editing with Jin Billy Li.]
Thank you to Genomes Unzipped for giving us the opportunity to write about our paper published in Nature Methods [1]. Our goal was to develop a method to identify RNA editing sites using matched DNA and RNA sequencing of the same sample. Looking at the problem initially, it seems straightforward enough to generate a list of variants using the RNA sequencing data and then filter out any variants that also appear in the DNA sequencing. In reality, one must pay close attention to the technical details in order to discern true RNA editing sites from false positives. In this post, we will highlight a couple of key strategies we employed to accurately identify editing sites.
Background
Last year in Science, Li et al. published a study where they called human RNA editing sites using matched DNA and RNA sequencing [2]. They reported over 10,000 RNA-DNA-Differences (RDDs) of all twelve mismatch types, many of which were predicted to introduce amino acid changes in protein sequences. This result was quite surprising in the light that only two known mechanisms exist for RNA editing in humans: adenosine to inosine (interpreted during translation as guanosine) deamination (A-to-I editing) and the much less common cytosine to uracil deamination (C-to-U editing). If true, this result would imply the existence of multiple unknown mechanisms that act to modify RNA transcripts. As many of you who follow this blog already know, some issues were raised about the validity of these RDD sites. Three technical comments were published by Science showing that almost all RDD sites found by Li et al. are false positives (full disclosure: one of the comments was written by us). A more thorough discussion about these technical comments can be found here.
In contrast to the results presented by Li et al., two subsequent studies by Bahn et al. [3] and Peng et al. [4] found that the majority of human RNA editing sites are of the A-to-I type and that very few of these sites introduce amino acid changes (although it’s not fair to compare these two studies with Li et al., as explained later on in this post). Interestingly, these two studies were able to validate a few non-A-to-I editing sites using PCR coupled with Sanger sequencing. This would seem to suggest that these novel non-canonical editing mechanisms do exist, albeit acting at drastically fewer sites than reported by Li et al. In our opinion, the evidence for these non-canonical editing sites was not entirely convincing, because the validation rate for them was much lower than the A-to-I editing sites. It seemed entirely possible to us that the small number of validated non-canonical editing sites were technical artifacts (it wouldn’t be genomics without some false positives!). As we developed our method, the big question looming in the back of our minds was whether we would find additional evidence to support these novel editing types.
Mapping strategy for RNA-seq reads
A pivotal question we dealt with was how to map our RNA-seq reads. There exist many software packages and pipelines that can be used to make RNA-seq alignments. Like many other problems in bioinformatics, if you ask ten different people you will get ten different answers. Should we map our reads to the genome or to the transcriptome? Which alignment algorithm should we use?
For our goal (variant calling) we knew that we had to make as accurate of an alignment as possible. Keeping this in mind, let’s take a look at the first question: genome or transcriptome alignment? The major advantage of alignment to the transcriptome is that sequencing reads spanning exon-exon splicing junctions have no trouble finding their correct alignment, but (and this is crucial) the major disadvantage is that transcriptome annotations are incomplete. If sequencing reads that originate from unannotated transcripts are forced onto the transcriptome, they may align spuriously to the wrong transcript. This would cause false mismatches. Therefore we decided to align our sequencing reads to the genome. But how do we deal with reads that span exon-exon junctions? If we align our reads to the genome only, we will encounter situations as illustrated in Figure 1a; a sequencing read originating from an exon-exon junction can be incorrectly mapped to a paralogous gene with a mismatch. To avoid this problem, we mapped our reads simultaneously to the genome and sequences spanning all known exon-exon junctions as illustrated in Figure 1b.
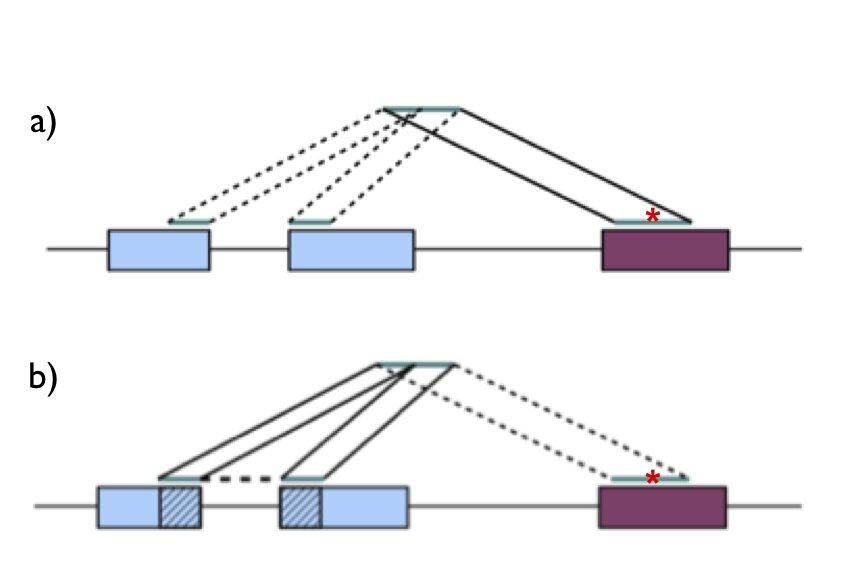
Figure 1-Alignment of reads spanning exon-exon junctions. In both panels, the blue boxes represent a gene from which an RNA sequencing read is derived, and the maroon box represents a paralog. The correct alignment is across the splice junction in the blue gene, while the read can also align to the maroon gene with a mismatch (red asterisk). a) Mapping of a junction-spanning read to the genome only can cause a spurious alignment with mismatch to a paralogous gene. b) Mapping of a junction-spanning read to genome and exon-exon junction sequences rescues the correct alignment.
The other choice to make was which alignment software to use. Alignment algorithms fall into two major categories: ones that allow alignment with gaps or ones that do not. An ungapped aligner (such as Bowtie) will align reads much faster than a gapped aligner, but will not provide accurate alignments around insertion or deletion polymorphisms. These inaccuracies may be inconsequential when using RNA-seq to measure gene expression levels, but for variant calling we want to avoid these false mismatches introduced at indel sites. Therefore we chose to use a gapped alignment algorithm (BWA).
Evaluating performance separately in Alu and non-Alu regions of the genome
It is well established that a large number of human A-to-I editing sites (hundreds of thousands, if not more) lie inside Alu repeats. The reason for this is that adenosine deaminase enzymes act on double-stranded RNA substrates. In many genes, Alu repeats frequently occur in pairs with inverse orientations that easily form dsRNA structures. When looking for RNA editing sites in the human genome, the vast majority of sites found will be A-to-I editing sites in Alu repeats. This is the reason why it’s not fair to compare editing sites from Li et al. (who only focus on non-repetitive regions of the genome) with Bahn et al. and Peng et al. (who both call editing sites genome-wide). Naturally the studies that look for editing sites genome-wide will have a higher enrichment of A-to-I editing sites (mostly in Alus) than studies that only look for editing sites in non-repetitive regions of the genome. We want to stress that a more informative metric is to examine editing sites outside of Alu repeats where editing is much less frequent and therefore substantially harder to accurately detect. Indeed we found that to accurately detect editing sites outside of Alu repeats, we had to include filters to remove false positives caused by read mapping issues (misalignment at microsatellite repeats, homopolymer runs, splicing junctions, and paralogs).
Our results are heavily enriched with A-to-I editing sites
The most important point from our results can be seen in Figure 2. Although we designed filters that were unbiased toward detecting A-to-I editing, we found that almost all of our called editing sites are indicative of A-to-I editing (A-to-G mismatch type). We compared our editing sites to the ones discovered in the previous studies mentioned above (Table 1 in the paper). Unsurprisingly, for editing sites in Alu repeats, all studies found a substantial enrichment of A-to-G mismatches. On the other hand, for editing sites that are not in repetitive elements our method finds a substantially higher fraction of A-to-G mismatches. We find 87% of our non-repetitive editing sites are A-to-G mismatches, while the other studies have A-to-G fractions of less than 50% for their non-repetitive sites.
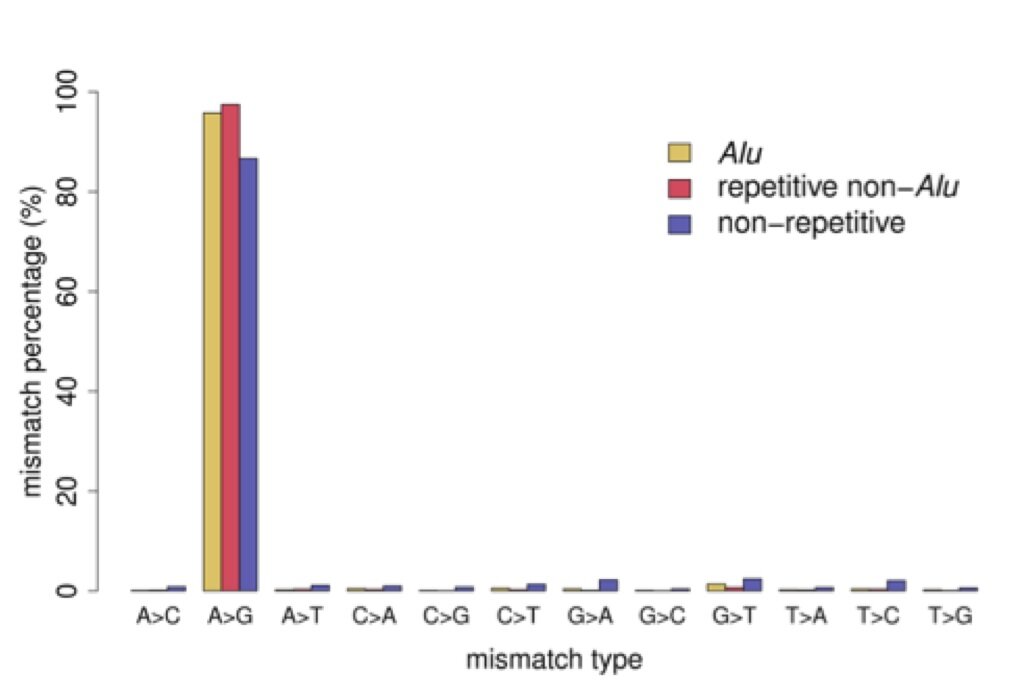
Figure 2 – Percentage of all twelve mismatch types. Mismatch sites were separated into 3 sets: those in Alu repeats, those in other repeats (non-Alu), and those not in any repetitive element
So, what’s the deal with these non-A-to-I sites?
We believe that most if not all of the non-A-to-G mismatches are caused by technical artifacts for two reasons. Firstly, we were unable to validate any of these non-A-to-G sites using PCR and Sanger sequencing. We were careful to design PCR primers that only amplified the gene of interest. If PCR primers are not carefully designed, they can amplify a paralogous gene in addition to the desired gene. This can cause a double peak in the cDNA sequencing trace which could then be falsely interpreted as a “validation” signal. Secondly, we observed that after each of our filtering steps, the ratio of A-to-G sites to non-A-to-G sites increased (Supplementary Figure 2 in the paper). This strongly suggests that for every filter we impose, the non-A-to-G mismatches are the false positives being filtered away.
Future Directions
The development of high-throughput sequencing technologies has resulted in the identification of numerous A-to-I RNA editing sites. However, identification of editing sites is only the initial step towards understanding the biological significance of RNA editing. We look forward to future studies that identify which editing sites are functional and further explore their role in both cellular and organismal contexts.
—
[1] Ramaswami et al. (2012) Accurate identification of human Alu and non-Alu RNA editing sites. doi:10.1038/nmeth.1982.
[2] Li et al. (2011) Widespread RNA and DNA Sequence Differences in the Human Transcriptome. doi: 10.1126/science.1207018.
[3] Bahn et al. (2012) Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. doi: 10.1101/gr.124107.111.
[4] Peng et al. (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. doi:10.1038/nbt.2122.








 RSS
RSS Twitter
Twitter
I’m curious – I just had a look over your paper and there are sites you’ve found (quite a lot of them in fact) that exhibit 100% rates of editing, where the nucleotide is consistently different in the RNA. Any comments on this?
I can see how a mechanism could evolve where it is useful on occasion to change the base encoded, depending on circumstances, in a similar fashion to the use of alternative splice variants. i.e. these single edits might have some functional significance which are useful during certain physiological states. But 100% editing seems kind of weird.
Obviously, biological is weird, and also you’ve only got one time point (perhaps another sample from the same individual would show different rates of editing at another time point depending on physiological state?), but this struck me as quite unusual.
Nice post. Enjoyed reading. This has been a week/month of RNA Editing with so many interesting papers highlighting the next-gen sequencing artifacts.
Here is another interesting paper showing the non-canonical RNA Editing sites are artifact, but in mouse.
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0033720#pone.0033720-Bahn1
The paper could also find the split of artifacts causing the edits.
“the strand bias artifacts account for roughly 70% of observed non-canonical editing sites, reference genome errors account for 15% of artifacts, and the remaining 15% are likely the product of other reference genome imperfections”
Just a small clarification. I think the “maroon box” in the Figure 1 is not just any paralog, but a pseudo-gene with no introns. Because legit paralogs have the same intron-exon structure.
I think that is among the such a lot vital information for me. And i’m happy studying your article. But wanna commentary on some common things, The web site style is ideal, the articles is actually nice : D. Just right job, cheers
I just wonder. If you use splice-aware program like Tophat, can you just map your read to genome instead of map to both annotate transcript and genome.