 Now the dust has settled, I’ve been reflecting on the controversial recommendation from the American College of Medical Genetics and Genomics (ACMG) that all clinical genomes should be screened for a specific set of conditions. Following the release of the guidelines, the European Society of Human Genetics (ESHG) published its more conservative recommendations, and vigorous debate has continued internationally regarding the wisdom of introducing genomic screening. While I still have some major reservations about the policy (outlined in previous posts), upon reflection there are certainly things some aspects that make a lot of sense…
Now the dust has settled, I’ve been reflecting on the controversial recommendation from the American College of Medical Genetics and Genomics (ACMG) that all clinical genomes should be screened for a specific set of conditions. Following the release of the guidelines, the European Society of Human Genetics (ESHG) published its more conservative recommendations, and vigorous debate has continued internationally regarding the wisdom of introducing genomic screening. While I still have some major reservations about the policy (outlined in previous posts), upon reflection there are certainly things some aspects that make a lot of sense…
Tag Archive for 'incidental findings'
 The ongoing debate about whether, what, when and how to feedback incidental findings (IFs) from whole genome sequencing continues to rage on both sides of the Atlantic following the American College of Medical Genetics and Genomics’ controversial recommendations on reporting IFs released last month. In an unexpected twist, the authors of the guidance have now written “a clarification” in response to the many criticisms that have been raised including here on GenomesUnzipped. The clarification covers five points – autonomy, children, labs, communication and interpretation.
The ongoing debate about whether, what, when and how to feedback incidental findings (IFs) from whole genome sequencing continues to rage on both sides of the Atlantic following the American College of Medical Genetics and Genomics’ controversial recommendations on reporting IFs released last month. In an unexpected twist, the authors of the guidance have now written “a clarification” in response to the many criticisms that have been raised including here on GenomesUnzipped. The clarification covers five points – autonomy, children, labs, communication and interpretation.
Continue reading ‘ACMG guidelines on IFs – responding to the response…’
Guest Co-Author: Dr Anna Middleton is an Ethics Researcher and Registered Genetic Counsellor, based at the Wellcome Trust Sanger Institute, UK.
 The American College of Medical Genetics (ACMG) has recently published recommendations for reporting incidental findings (IFs) in clinical exome and genome sequencing. These advocate actively searching for a set of specific IFs unrelated to the condition under study. For example, a two year old child may have her (and her parents’) exome sequenced to explore a diagnosis for intellectual disability and at the same time will be tested for a series of cancer and cardiac genetic variants. The ACMG feel it is unethical not to look for a series of incidental conditions while the genome is being interrogated, conditions that the patient or their family may be able to take steps to prevent. This flies in the face of multiple International guidelines that advise against testing children for adult onset conditions. The ACMG justify this as “a fiduciary duty to prevent harm by warning patients and their families”. They conclude that “this principle supersedes concerns about autonomy”, i.e. the duty of the clinician to perform opportunistic screening outweighs the patients right not to know about other genetic conditions and their right to be able to make autonomous decisions about testing.
The American College of Medical Genetics (ACMG) has recently published recommendations for reporting incidental findings (IFs) in clinical exome and genome sequencing. These advocate actively searching for a set of specific IFs unrelated to the condition under study. For example, a two year old child may have her (and her parents’) exome sequenced to explore a diagnosis for intellectual disability and at the same time will be tested for a series of cancer and cardiac genetic variants. The ACMG feel it is unethical not to look for a series of incidental conditions while the genome is being interrogated, conditions that the patient or their family may be able to take steps to prevent. This flies in the face of multiple International guidelines that advise against testing children for adult onset conditions. The ACMG justify this as “a fiduciary duty to prevent harm by warning patients and their families”. They conclude that “this principle supersedes concerns about autonomy”, i.e. the duty of the clinician to perform opportunistic screening outweighs the patients right not to know about other genetic conditions and their right to be able to make autonomous decisions about testing.
One of the major bioethical debates in clinical genetics and genomics research is the issue of what to do with incidental or secondary findings (IFs) unrelated to the original clinical or research question. Every genome contains thousands of rare variants, including a surprising number of loss of function variants, as well as hundreds of variants associated with common disease and dozens linked with recessive conditions. As whole genome or exome sequencing is used more routinely in non-anonymised cohorts – such as the 100,000 patient genomes to be sequenced by the UK NHS – these variants will be uncovered and linked to an increasing number of individuals. What should we do with them?
Robert Green of Brigham and Women’s Hospital in Boston, who co-chairs the American College of Medical Genetics (ACMG) working group on secondary findings, was quoted in a Nature blog last year saying, “we don’t think it’s going to be a sustainable strategy for the evolving practice of genomic medicine to ignore secondary findings of medical importance”. But just saying it doesn’t make it so. There are still numerous questions that need to be addressed – you can be part of the debate by participating in the Sanger Institute’s Genomethics survey.

Gholson Lyon is a physician-scientist currently working at the Utah Foundation for Biomedical Research and the Center for Applied Genomics at Children’s Hospital of Philadelphia. He will be starting as an assistant professor in human genetics at Cold Spring Harbor Laboratory next month. I asked him to write this guest post to provide some personal context to his thought-provoking commentary in Nature (subscription required) on returning genetic findings to research subjects. [DM]
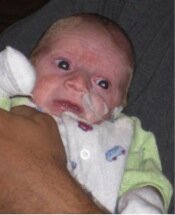
Photo of Max, who died aged four months from Ogden syndrome. Posted with permission from his family.
I have just published in Nature a commentary discussing the need to bring exome and genome sequencing into the clinical arena, so that these data are generated with the same rigorous clinical standards as for any other clinical test. This way, we can then easily return at least medically actionable results to research participants. In this day and age of consumer and patient empowerment, I can also see eventually returning all data, including the raw data, to any interested participants, as this can then promote crowd-sourcing for data analysis, with research participants controlling and promoting the relative privacy of and analysis of their own data.
As I described in my commentary, my thinking on this matter was prompted mainly by Max (see picture) and his family. The obituary for Max can be found here, and that of his cousin, Sutter, here. We described their condition here, and we named this new disease Ogden Syndrome in honor of where the first family lives. I am now trying to think about and discuss the human aspects of and lessons from this story. My thinking has also been influenced somewhat by the late James Neel, who wrote a very thought-provoking book called Physician to the Gene Pool.
To me, it was deeply disconcerting that I could not officially return any results to this family (or to another family in a different project discussed here) even when the papers describing the genetic basis of their disease were published, as this was considered “research” and was not performed in a clinically appropriate (CLIA-certified) manner. This was all the more painful when one of the sisters in the Ogden family became pregnant and asked me what I knew. I cannot predict whether it would have helped or hurt this woman to learn during her pregnancy that she was indeed a carrier of the mutation, with the associated 50% risk of her baby boy having the disease. I also do not know if she would have undergone any genetic testing via amniocentesis of the fetus prior to birth (with the associated ~1% risk of miscarriage from the procedure), nor do I know what decisions she might have made prior to the birth even if she had undergone such testing. All in all, it was certainly an ethical and moral dilemma for me not to be able to return the research result to her, given that the results were not obtained in a CLIA-certified manner. It is still an issue, as there are even now financial and systematic barriers for getting all women in the family tested with a CLIA-certified gene test for NAA10 (which was developed over a six month period by ARUP Laboratories). It would have been so much better if we had just done the entire sequencing up front in a CLIA-certified manner.
Continue reading ‘Guest post: Time to bring human genome sequencing into the clinic’
 RSS
RSS Twitter
Twitter
Recent Comments