Over the last several years, the number of genetic variants unambiguously associated with disease risk has grown dramatically. However, interpreting these signals has been extremely difficult—most of the identified variants do not disrupt genes, and indeed many don’t fall anywhere near genes (this observation has even led some to discount these signals entirely). To an investigator interested in following up on these signals, this is somewhat depressing: how can we hope to explore how polymorphisms affect disease risk if they don’t seem to fall in any sort of genome annotation that we understand?
In this context, I thought I’d point to an important paper that, among many other things, gives the first systematic evidence that variants which influence disease are not just randomly scattered across the genome, but instead tend to fall in particular regions—in particular, enhancer elements (regions where DNA-binding proteins interact with DNA to influence gene expression).
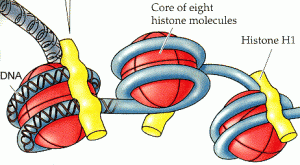
The authors rely on the fact that, in the cell, DNA is wrapped around proteins called histones, which control how accessible the DNA is to things like transcription factors (see above figure). These proteins can be chemically modified, and it is now clear that particular patterns of modifications are predictive of the function of the DNA in the region—some modifications indicate transcribed genes, others regions of enhancer activity, others repressed regions, etc.
What the authors did in this study was generate genome-wide maps of several histone modifications in nine different cell types, and use this data to predict the function of each 200 base pair segment of the human genome in each cell type. There are a number of interesting analyses of these “maps” of genome function in the paper, but for our purposes here there’s one of particular interest: the authors took sets of SNPs associated with various diseases and simply asked, are these variants enriched in regions with any particular functional prediction? And indeed, for several phenotypes, there is a striking enrichment of association signals in enhancers elements in a relevant cell type. For example, SNPs which influence lipid levels are enriched in enhancers in a liver cancer cell line, and SNPs which influence the autoimmune disease lupus are enriched in enhancers in a lymphoblastoid cell line.
As these types of functional maps are generated in more cell types, I imagine there will be more stories like this. The problem with interpreting disease association studies, it seems likely, is largely due to our lack of understanding of genome function.
—-
Citation: Ernst et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. doi:10.1038/nature09906
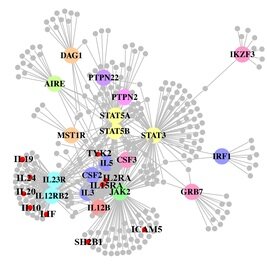 A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ] There is a real “wow” paper out in pre-print at the journal Genetics in Medicine. It is a wonderful example of the application of cutting edge sequencing technology to solve a medical mystery. Even better, the authors also include an auxiliary discussion about the medical and ethical issues surrounding the diagnosis, which raises some interesting issues about the transition from research to clinical sequencing.
There is a real “wow” paper out in pre-print at the journal Genetics in Medicine. It is a wonderful example of the application of cutting edge sequencing technology to solve a medical mystery. Even better, the authors also include an auxiliary discussion about the medical and ethical issues surrounding the diagnosis, which raises some interesting issues about the transition from research to clinical sequencing. RSS
RSS Twitter
Twitter
Recent Comments