This guest post was contributed by Joseph Buxbaum, Mark Daly, Silvia De Rubeis, Bernie Devlin, Kathryn Roeder, and Kaitlin Samocha from the Autism Sequencing Consortium (see affiliations and details at the end of the post).
Autism spectrum disorder (ASD) is a highly heritable condition characterized by deficits in social communication, and by the presence of repetitive behaviors and/or stereotyped interests. While it is clear from family and twin studies that genetic factors contribute strongly to the onset of this disorder, the search for specific risk genes for ASD has only recently begun to yield fruit. Finding these specific genes is critical not only for providing potential diagnoses for individual families, but also for obtaining insights into the pathological processes that underlie this neurodevelopmental disorder, which may ultimately lead to novel therapeutic approaches. Identification of ASD genes may at some point also reveal part of what makes us social beings.
In a paper published in Nature last week we and the other members of the Autism Sequencing Consortium (ASC) describe the application of whole exome sequencing (WES), selectively sequencing the coding regions of the genome, to identify rare genetic variants and then genes associated with risk for ASD. WES data were analyzed from nearly 4,000 individuals with autism and nearly 10,000 controls. In these analyses, we identify and subsequently analyze a set of 107 autosomal genes with a false discovery rate (FDR) of <30%; in total, this larger set of genes harbor de novo loss of function (LoF) mutations in 5% of cases, and numerous de novo missense and inherited LoF mutations in additional cases. Three critical pathways contributing to ASD were identified: chromatin remodelling, transcription and splicing, and synaptic function. Chromatin remodelling controls events underlying neural connectivity. Risk variation also impacted multiple components of synaptic networks. Because a wide set of synaptic genes is disrupted in ASD, it seems reasonable to suggest that altered chromatin dynamics and transcription, induced by disruption of relevant genes, leads to impaired synaptic function as well.
In this post we wanted to focus on an easily-overlooked aspect of this paper: the use of a false discovery rate (FDR) approach to identifying genes for follow-up analysis. While FDR is a well-recognized approach in biology, one could also argue for using a family wise error rate (FWER), which has been the norm in recent large-scale, genome-wide association studies (GWAS). So why did we decide to take this alternative approach here?
Continue reading ‘Incorporating false discovery rates into genetic association in autism’
 As I
As I  Out in Nature this week is a paper by three Genomes Unzipped authors reporting 71 new genetic associations with inflammatory bowel disease (IBD). This breaks the record for the largest number of associations for any common disease, and includes many new and interesting biological insights that you should all go and read about in the paper itself (pay-to-access I’m afraid) or on the Sanger Institute’s website.
Out in Nature this week is a paper by three Genomes Unzipped authors reporting 71 new genetic associations with inflammatory bowel disease (IBD). This breaks the record for the largest number of associations for any common disease, and includes many new and interesting biological insights that you should all go and read about in the paper itself (pay-to-access I’m afraid) or on the Sanger Institute’s website.
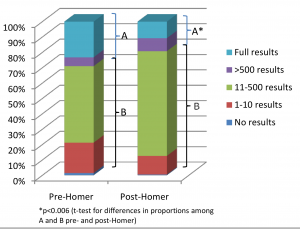 In PLoS Genetics this week there is a viewpoint article on data sharing in disease genetics. The authors systematically looked at 643 genome-wide association studies published between 2002 and 2010, to see how easily available the results of the studies are now. They found that the availability of full study results has gone down over time, and many groups that do share data have put more restrictions in place on its use. They put this down to fears over the privacy of research subjects, and in particular to the Homer et al study. The Homer et al result is somewhat complicated, but in essence it says that if you have stolen someone’s genotype data, you can use it to figure out if they have participated in any given research study by looking at the full results of the study.
In PLoS Genetics this week there is a viewpoint article on data sharing in disease genetics. The authors systematically looked at 643 genome-wide association studies published between 2002 and 2010, to see how easily available the results of the studies are now. They found that the availability of full study results has gone down over time, and many groups that do share data have put more restrictions in place on its use. They put this down to fears over the privacy of research subjects, and in particular to the Homer et al study. The Homer et al result is somewhat complicated, but in essence it says that if you have stolen someone’s genotype data, you can use it to figure out if they have participated in any given research study by looking at the full results of the study. Genome-wide association studies have been hugely successful in identifying dozens of common genetic risk factors for a large number of common diseases. However, one area that GWAS has not had much success in is the field of psychiatric illness, where finding common risk factors that replicate across studies has been consistently difficult. However, it looks like this is starting to change. The current issue of Nature Genetics has two papers from the Psychiatric GWAS Consortium, detailing some of the largest meta-analyses of schizophrenia and
Genome-wide association studies have been hugely successful in identifying dozens of common genetic risk factors for a large number of common diseases. However, one area that GWAS has not had much success in is the field of psychiatric illness, where finding common risk factors that replicate across studies has been consistently difficult. However, it looks like this is starting to change. The current issue of Nature Genetics has two papers from the Psychiatric GWAS Consortium, detailing some of the largest meta-analyses of schizophrenia and 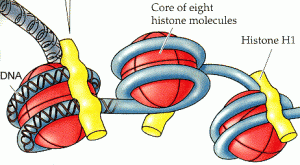
 RSS
RSS Twitter
Twitter
Recent Comments