 This guest post was contributed by Karol Estrada, a postdoctoral research fellow in the Analytic and Translational Research Unit at Massachusetts General Hospital and the Broad Institute of MIT and Harvard. It is dedicated to the memory of Laura Riba.
This guest post was contributed by Karol Estrada, a postdoctoral research fellow in the Analytic and Translational Research Unit at Massachusetts General Hospital and the Broad Institute of MIT and Harvard. It is dedicated to the memory of Laura Riba.
Genome-wide association studies (GWAS) of common variants have successfully implicated more than 70 genomic regions in type 2 diabetes, revealing new biological pathways and potential drug targets. However, most large studies have examined genetic variation only in northwestern European populations, despite the rich genetic diversity in other populations around the world. Most studies have also been limited in their ability to detect variants present in fewer than 5 percent of people. Much remains to be learned.
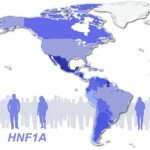
In this post, we discuss our new paper, published in the Journal of the American Medical Association, on a low-frequency missense variant in the gene HNF1A that raises risk of type 2 diabetes five-fold, and was seen only in Latinos. This variant was the only rare variant to reach genome-wide significance in an exome sequencing study of almost 4,000 people, the largest such study to date. We explain the ramifications for sample sizes of rare-variant studies, note the importance of studying populations outside of northwestern Europe, and caution against simplistic dichotomous interpretations of disease as either complex or monogenic. Finally, we note that a low-frequency or rare variant might guide therapeutic modification.
Continue reading ‘A rare variant in Mexico with far-reaching implications’
 This is a guest post by Peter Cheng and Eliana Hechter from the University of California, Berkeley.
This is a guest post by Peter Cheng and Eliana Hechter from the University of California, Berkeley.






 But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none.
But last spring Daniel alerted me to the 23andMe “DNA Day” sale. It was affordable, and at that point enough of the readers of my weblog had been typed that I kept getting questions as to my own background (e.g., my family has the title Khan, so there was a question as to whether I carried the “Genghis Khan haplotype”). So I bit. At the time I recall emailing Dan and being excited that I’d be told I likely had brown eyes and was 75% “European” and 25% “Asian.” When my results came back, I was in for a mild surprise. The proportion to the left are calculated by 23andMe’s “ancestry painting” algorithm. As you can see, I’m more than 25% “Asian.” My initial reaction was that this seemed a touch high, but no worries, I would ask around and see which other South Asians had such a high value. After dozens of instances of “gene sharing,” the answer came back: none. RSS
RSS Twitter
Twitter
Recent Comments