In the recent report from the US Government Accountability Office on direct-to-consumer genetic tests, much was made of the fact that risk predictions from DTC genetic tests may not be applicable to individuals from all ethnic groups. This observation was not new to the report – it has been commented on by numerous critics ever since the inception of the personal genomics industry.
So, why does risk prediction accuracy vary between individuals and what can be done to combat this? Are the DTC companies really to blame?
To explore these questions it is first necessary to understand what is meant by the odds ratio (OR). In genetic case-control association studies the OR typically represents the ratio of the odds of disease if allele A is carried compared to if allele B is carried. If all else is equal, genetic loci with a higher OR are more informative for disease prediction – so getting an accurate estimate is extremely important if prediction underpins your business model. However, getting an accurate estimate of OR is far from easy because many, often unmeasured, factors can cause OR estimates to vary. In this post I will try to break down the concept of a single, fixed odds ratio for a disease association, and highlight a number of factors that can cause odds ratios to vary using examples from the scientific literature.
The environmentally unfriendly odds ratio
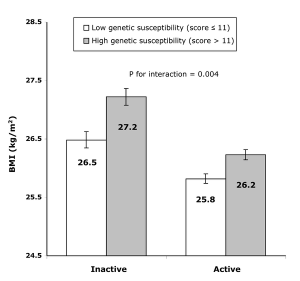 In a recent ‘Friday Links’ Luke highlighted a paper in PLoS Medicine showing physical activity can modifiy ORs of BMI associated loci. This is broadly termed ‘gene-environment interaction’ and until recently was not very widely investigated. I agree with Larry Parnell’s comment in response to Luke’s post that genetic studies must increasingly account for environmental factors.
In a recent ‘Friday Links’ Luke highlighted a paper in PLoS Medicine showing physical activity can modifiy ORs of BMI associated loci. This is broadly termed ‘gene-environment interaction’ and until recently was not very widely investigated. I agree with Larry Parnell’s comment in response to Luke’s post that genetic studies must increasingly account for environmental factors.
One question I often get asked after giving a presentation on the genetics of autoimmune disease goes something like this:
Autoimmune diseases have increased in frequency a great deal over the past 100 years and obviously the frequency of risk variants cannot have increased at the same rate – surely this indicates that autoimmune disease are caused by environmental rather than genetic factors.
And so I become embroiled, once again, in the age-old Nature vs Nurture debate. As Matt Ridley would say, what is clearly going on is Nature via Nurture. If the OR of a genetic variant increases over time the associated disease can increase in frequency, and a change of environment is one means of doing this. The most widely supported hypothesis for the pathogenesis of Crohn’s disease is an abnormal immune response to ordinary gut bacteria in genetically susceptible individuals. The environmental component, the gut bacteria, has changed substantially with increasing Westernisation and perhaps one consequence of this is that variants that previously had little or no effect on Crohn’s disease risk now have a much greater impact (even though they have not substantially changed frequency).
This obviously presents a problem for DTC companies because they must use odds ratio estimates to predict disease risk without considering the environment in which customers reside. The size of the problem therefore really hinges on how much odds ratio estimates vary between different environments and the scientific community has, to date, put little effort into understanding gene-environment interaction.
The travel-sick odds ratio
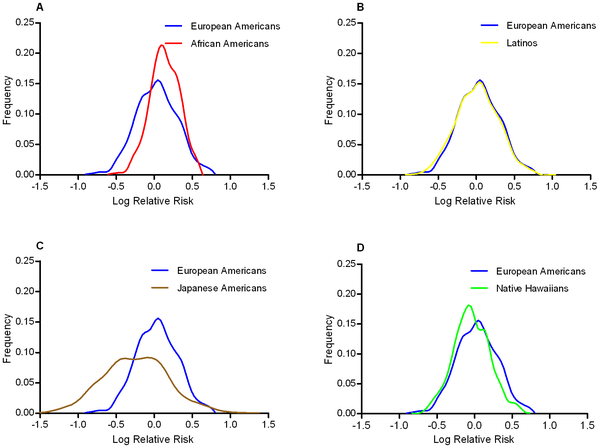
If we should be concerned about the effect of the environment on OR estimates then we should be very concerned indeed about taking OR estimates from European samples and applying them to risk prediction algorithms for Asian or African individuals. Across populations it is not only environments that differ but also things like allele frequencies and linkage disequilibrium patterns, and these can all have an effect on ORs. Somewhat reassuringly, a recent study published in PLoS Genetics showed that odds ratios at 19 Type II diabetes associated SNPs were consistent across European Americans, African Americans, Latinos, Japanese Americans, and Native Hawaiians.
OR consistency across continental groups is not always reported. In a manuscript that is currently an advanced online publication at Nature Genetics, an international group of scientists report a genomic region underlying susceptibility to glaucoma, the leading cause of irreversible blindness worldwide. In their European samples the odds ratio of the most associated genetic variant was 1.27. However, in a small sample of Asian individuals the odds ratio was an almighty 3.33. Kari Stefansson from Decode Genetics, who led the study, said
What this basically means is that when you’re developing diagnostics, you have to be aware of the potential geographic differences. You have to map them out so you can report different results in different populations.
Is this just an extreme case of gene-environment interaction or is something even more complex going on? As usual, the devil is in the details. In Europeans, the risk associated allele has a frequency of 20-30% but in the small number of Asian samples they genotyped it has a frequency of 0.3-0.6%. A consequence of the small sample size and low allele frequency is that the 95% confidence interval around the Asian OR estimate is 1.56-7.08, a big range in anybody’s book. So while I agree with Stefansson’s words regarding accounting for population differences in OR estimates, I also think it is important that large samples sizes are used to enable accurate OR estimates to be used in predictive algorithms.
DTC companies seem to be aware of the problem of applying point OR estimates across different ethnicities. 23andMe currently allow you to select your ethnicity prior to estimating your disease risk, but for the vast majority of diseases only a European option is available. This is really no fault of the DTC companies as very few studies have estimated ORs using large ethnically diverse sample sets and 23andMe provide a warning to this effect.
While we at 23andMe do actively look for associations that have been repeated in populations other than the original study population, we can only report data published by the scientific community at large. We feel it would be inappropriate to assume that studies performed in populations of a particular ethnic composition apply to everyone.
Age concern
Odds ratio’s can also vary according to age. A study published in this month’s edition of the New England Journal of Medicine by the GABRIEL consortium reports a risk locus that appears to increase risk of early-onset Asthma (onset before 16 years) but has little or no effect on adult-onset Asthma. However, incorporating such findings into predictive algorithms is not straightforward because one needs to carefully account for variation in OR throughout aging. The high-risk conveyed by the associated allele does not suddenly fall away on the morning of one’s sixteenth birthday. Very few scientific studies have attempted to estimate how ORs correlate with age, so it is also currently unrealistic to expect DTC companies to incorporate this information in their prediction algortihms.
Again, 23andMe seem aware of the problem of age confounding OR estimates because they provide a means of varying age before disease risk is calculated. However, if one plays around with this tool it is clear that, for the vast majority of diseases, little actually changes with respect to disease risk. Again, I don’t think this is the fault of 23andMe but rather reflects the little amount of scientific endeavour in this area.
Summary
I have highlighted a few confounding factors that can cause ORs to vary. Initial investigations seem to suggest that each of these will only have a small impact on OR estimates, though further investigation is certainly warranted and taken together the effects could be larger. The good news is that DTC companies seem to be aware of these issues and already have mechanisms in place to account for these confounding factors when scientific research allows them to accurately do so.
It is currently unrealistic to expect DTC companies to account fully for environmental, population and age differences in their prediction algorithms. We scientists need to first do the research that will allow potential confounding factors to be accounted for in prediction algorithms. Only through studying large prospective cohorts will these confounding factors be fully characterised and we must extend our research to groups of non-European ancestry.








 RSS
RSS Twitter
Twitter
Congratulations to all the contributors for choosing always interesting topics. This time we deal with gene x environment, a pair that we should never forget when we read about a new genetic association. I find particularly interesting the ethnicity issue. In a group a SNP has a high risk, in another the risk is low: of course this happens because there are other variants involved, variants that are found only in one of the two groups. Maybe one day we will know more about genetic variation among populations, and we’ll be able to predict the real risk regardless of ethnicity.
Nice post! Larry is right and we beat the same drum usually so it’s good to see the GxE issue explored. It’s probably going to be essential to tease out as much as possible – just a random example that I was reading when I read the post: http://bit.ly/a7jPL0 Diabetes is associated with pollution (causative? or urban lifestyle?) which obviously is linked to where you live. Next it would be nice to see if there are specific genes that interact with pollution and affect the risk. The benefits of knowing as much as possible about both G and E include a) more precise prediction and most importantly b) the E part can be changed to reduce the risk.
One post I would like to see here is the explanation of OR and RR, how they are related and why OR does not actually = risk. In some cases (HLA genes and Celiac) you can end up with ORs of 50-100 but these don’t translate to similar RR or absolute risk. Since this was perhaps one of the more valid criticisms of DTC it would be good to have it explained –