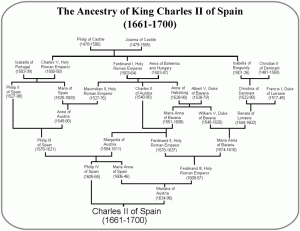
 Today is, of course, the day of the Royal Wedding, with new blood entering the British royal line, and the hope of new heirs to our throne. And of course the question on the lips of all British geneticists is: will there be any new royal genetic diseases in this crop? The European royal lines have always been prone to the odd loss-of-function mutation. An unlucky mutation in Queen Victoria’s Factor IX gene caused a nasty case X-linked Haemophilia B in her male descendants (a mutation that was only mapped in 2009 by sequencing the bones of the murdered Romanov branch). Luckily for them, this mutation hasn’t been observed in any of Victoria’s descendants lately; while it can hide undetected in women, this obviously doesn’t apply to William. More systemic genetic problems have been the result of heavy inbreeding; Charles II of Spain, with his distressingly bushy family tree (left), suffered from severe Habsburg jaw, along with a host of other genetic complaints.
Today is, of course, the day of the Royal Wedding, with new blood entering the British royal line, and the hope of new heirs to our throne. And of course the question on the lips of all British geneticists is: will there be any new royal genetic diseases in this crop? The European royal lines have always been prone to the odd loss-of-function mutation. An unlucky mutation in Queen Victoria’s Factor IX gene caused a nasty case X-linked Haemophilia B in her male descendants (a mutation that was only mapped in 2009 by sequencing the bones of the murdered Romanov branch). Luckily for them, this mutation hasn’t been observed in any of Victoria’s descendants lately; while it can hide undetected in women, this obviously doesn’t apply to William. More systemic genetic problems have been the result of heavy inbreeding; Charles II of Spain, with his distressingly bushy family tree (left), suffered from severe Habsburg jaw, along with a host of other genetic complaints.
In terms of inbreeding, there has been a bunch of digging around in the press to find the closest common ancestor of William and Kate: Channel Four turned up fourteen and fifteenth cousinships, and the Daily Mail managed to find a eleventh cousinship. For comparison, William’s parents Diana and Charles were also 11th cousins, and the Queen and Prince Philip were a far more regal 2nd cousins once removed. Eleventh cousins share on average 60-parts-per-billion of DNA, or about 180bp (although with wide variation due to the spotty nature of meiotic recombination: in fact, 99.5% of 11th cousins will share no stretches of DNA through recent descent at all, while the remaining 0.5% will typically share tens of thousands of bases). Given that the average person harbours about 10 recessive diseases, this gives about a 1 in 1.6 million chance of Kate and Will’s offspring developing a royal disease due to a piece of DNA shared between them. So, not very likely then.
In fact, eleventh cousins is a pretty low degree of relatedness, by the standard of these things. A study of inbreeding in European populations found that couples from the UK are, on average, as genetically related as 6th cousins (the study looked at inbreeding in Scots, and in children of one Orkadian and one non-Orkadian. No English people, but I would be very suprised if we differed significantly). 6th cousins share about 0.006% of their DNA, and thus have about a 0.06% chance of developing a genetic disease via a common ancestor. Giving that the Royal Family are better than most at genealogy, we can probably conclude that the royal couple are less closely related than the average UK couple, and thus their children are less likely than most to suffer from a genetic disease. Good news for them, bad news for geneticists, perhaps?
 Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.
Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.
 I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ]
I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ]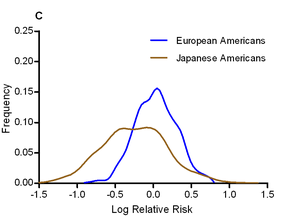 The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
 Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ]
Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ] Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say:
Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say: RSS
RSS Twitter
Twitter
Recent Comments