 Over at Nature News, Erika Check Hayden has a post about a recent Science Translational Medicine paper by Bert Vogelstein and colleages looking at the potential predictive power of genetics. The take-home message from the study (or at least the message that has been taken home by, e.g., this NYT article) is that DNA does not perfectly determine which disease or diseases you may get in the future. This take home message is true, and to me relatively obvious (in the same way that smoking doesn’t perfectly determine lung cancer, or body weight and dietary health doesn’t perfectly determine diabetes status).
Over at Nature News, Erika Check Hayden has a post about a recent Science Translational Medicine paper by Bert Vogelstein and colleages looking at the potential predictive power of genetics. The take-home message from the study (or at least the message that has been taken home by, e.g., this NYT article) is that DNA does not perfectly determine which disease or diseases you may get in the future. This take home message is true, and to me relatively obvious (in the same way that smoking doesn’t perfectly determine lung cancer, or body weight and dietary health doesn’t perfectly determine diabetes status).
A lot of researchers have had a pretty negative reaction to this paper (see Erika’s storify of the twitter coverage). There are lots of legitimate criticism (see Erika’s post for details), but to be honest I suspect that a lot of this is a mixture of indignation and sour grapes that this paper, a not particularly original or particularly well done attempt to answer a question that many other people have answered before, got so much press (including a feature in the NYT). A very large number of people have tried to quantify the potential predictive power of genetics for a number of years – why was there no news feature for me and Jeff, or David Clayton, or Naomi Wray and Peter Visccher, or any of the other large number of stat-gen folks who have been doing exactly these studies for years. ANGER RISING and so forth.
But of course, the reason is relatively obvious. Continue reading ‘Identical twins usually do not die from the same thing’

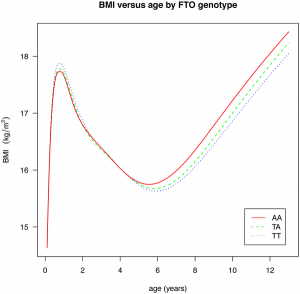 There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
 Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.
Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe. Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ]
Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ] Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say:
Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say: RSS
RSS Twitter
Twitter
Recent Comments