A lot of the Genomes Unzipped crew seem to be away on holiday at the moment, so today’s Links post may lack the the authorial diversity that you’re accustomed to.
I just got around to reading the August addition of PLoS Genetics, and found a valuable study from the Keck School of Medicine in California. They authors looked at the effect of known common variants in five American ethnic groups (European, African, Hawaiian, Latino and Japanese Americans), to assess how similar or different the effects sizes were across the groups.
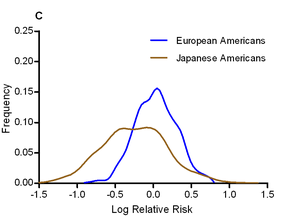 The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
The authors calculated odds ratios for each variant in each ethnic group, and looked for evidence of heterogeneity in odds ratios. They find that, in general, the odds ratios tend to show surprisingly little variation between ethnic groups; the direction of risk was the same in almost all cases, and the mean odds ratio was roughly equal across populations (the authors note that this pretty effectively shoots down David Goldstein’s “synthetic association” theory of common variation). One interesting exception was that the effect size of the known T2D variants was significantly larger in Japanese Americans, who had a mean odds ratio of 1.20, compared to 1.08-1.13 for other ethnic groups. The graph to the left shows the distribution of odds ratios in European and Japanese Americans.
These sorts of datasets will be very useful for personal genomics in the future, as a decade of European-centered genetics research has left non-Europeans somewhat in the lurch with regards to disease risk predictions. However, the problem with the approach in this paper is that even this in large a study (6k cases, 7k controls) the error bounds on the odds ratios within each group are still pretty large. [LJ]
Over at the Guardian Science Blog, Dorothy Bishop explains the difference between learning that a trait is heritable (e.g. from twin studies), and mapping a specific gene “for” a trait (e.g. via GWAS). Her conclusion is worth repeating:
The main message is that we need to be aware of the small effect of most individual genes on human traits. The idea that we can test for a single gene that causes musical talent, optimism or intelligence is just plain wrong. Even where reliable associations are found, they don’t correspond to the kind of major influences that we learned about in school biology. And we need to realise that twin studies, which consider the total effect of a person’s genetic makeup on a trait, often give very different results from molecular studies of individual genes.
There are also interesting questions to be asked about why there is such a gap between heritabilities estimated by twin studies, and the heritability that can be explained by GWAS results. That is, however, is a question for another day. [LJ]
Another article just released in PLoS Genetics provides a powerful illustration of just how routine whole-genome sequencing is now becoming for researchers: the authors report on complete, high-coverage genome sequence data for twenty individuals. The samples included 10 haemophilia patients and 10 controls, taken as part of a larger study looking at the genetic factors underlying resistance to HIV infection. While this is still a small sample size by the standards of modern genomics, there are a few interesting insights that can be gleaned from the data: for instance, the researchers argue from their data that each individual has complete inactivation of 165 protein-coding genes due to genetic variants predicted to disrupt gene function. I’ll be following up on this claim in a future post. [DM]

Finally, a quick shout-out to our fellow Sanger researchers, including Verneri Anttila and Aarno Palotie, along with everyone else in the International Headache Genetics Consortium, for finding the first robust genetic association to migrane. They looked at 3,279 cases and >10k controls (and another 3,202 cases to check their results), and found that the variant rs1835740 was significantly associated with the disease.
To tie in with the above story, in the region of 40-65% of variation in migraine is heritable, but only about 2% of this was explained by the rs1835740 variant. However, explaining heritability isn’t the main point of GWAS studies: a little follow-up found that rs1835740 was correlated with expression of the gene MTDH, which in turn suggests a defect in glutamate transport; hopefully this new discovery will help shed some light on the etiology of the disease. [LJ]








 RSS
RSS Twitter
Twitter
You might also notice that Genomes Unzipped’s very own Jeffrey C Barrett is on the migraine paper. :)
Meh, if we congratulated you every time your name appears on a GWAS paper we wouldn’t get much else done.
Note the incorrect interpretation of p-values in the Guardian piece:
Consider one of the more reliable associations between genes and behaviour: a gene known as KIAA0319 which has been found to relate to reading ability in several different samples. In one study, an overall association was reported with a p value of 0.0001, indicating that the likelihood of the association being a fluke is 1 in 10,000.
By coincidence, that PLoS Genetics paper on sequencing comes from the lab of… David Goldstein (synthetic association).
The figure of 165 homozygous protein-truncating variants per average genome seems awfully high. Indeed, the text indicates that the “real” number is probably rather lower, due to things like the inadvertent consideration of pseudogenes. But it would still be pretty high, say over 100. If there are 20,000 translated genes, that’s 1 in 200! The simplest cocktail-napkin calculation would then suggest that ~7% of translated autosomal genes are expressed by only one allele. Again, a non-intuitively high figure–especially considering the dosage effects of one-allele expression of some genes, famously BRCA1 and BRCA2.