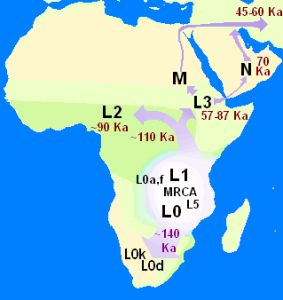
One thing we have done in Genomes Unzipped is to report on what is on the market for consumers interested in getting information about their genetic data. While we have found generally positive things to say about this market, there are also many exaggerated claims especially when it comes to making inferences about an individual’s ancestors from direct-to-consumer genetics companies. An example came up last summer with a BBC radio 4 interview of Alistair Moffat of Britain’s DNA. This post will discuss the scientific basis of some of the claims made in the interview.
But first of all, what is my motivation to write this post? After all, there are quite a few genetic ancestry companies like Britain’s DNA, making similar claims. Why specifically discuss this BBC radio 4 interview? The main reason is that listening to this radio interview prompted my UCL colleagues David Balding and Mark Thomas to ask questions to the Britain’s DNA scientific team; the questions have not been satisfactorily answered. Instead, a threat of legal action was issued by solicitors for Mr Moffat. Any type of legal threat is an ominous sign for an academic debate. This motivated me to point out some of the incorrect, or at the very least exaggerated, statements made in this interview. Importantly, while I received comments from several people for this post, the opinion presented here is entirely mine and does not involve any of my colleagues at Genomes Unzipped.
Continue reading ‘Exaggerations and errors in the promotion of genetic ancestry testing’





 RSS
RSS Twitter
Twitter
Recent Comments