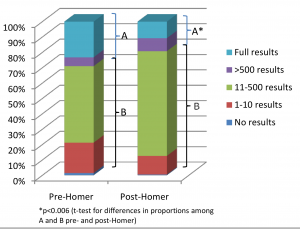
 In PLoS Genetics this week there is a viewpoint article on data sharing in disease genetics. The authors systematically looked at 643 genome-wide association studies published between 2002 and 2010, to see how easily available the results of the studies are now. They found that the availability of full study results has gone down over time, and many groups that do share data have put more restrictions in place on its use. They put this down to fears over the privacy of research subjects, and in particular to the Homer et al study. The Homer et al result is somewhat complicated, but in essence it says that if you have stolen someone’s genotype data, you can use it to figure out if they have participated in any given research study by looking at the full results of the study.
In PLoS Genetics this week there is a viewpoint article on data sharing in disease genetics. The authors systematically looked at 643 genome-wide association studies published between 2002 and 2010, to see how easily available the results of the studies are now. They found that the availability of full study results has gone down over time, and many groups that do share data have put more restrictions in place on its use. They put this down to fears over the privacy of research subjects, and in particular to the Homer et al study. The Homer et al result is somewhat complicated, but in essence it says that if you have stolen someone’s genotype data, you can use it to figure out if they have participated in any given research study by looking at the full results of the study.
It certainly seems possible that worries about privacy are reducing the free flow of information within the research community. However, whether on balance the decrease in information flow is worth the increase in security is an open question. For my own view, I feel that having the genome-wide results of genome-wide association studies freely available is very important to the field, and is more important than the the rather esoteric risk of someone stealing someone’s DNA and using it to figure out that they once took part in a research study of inflammatory bowel disease. [LJ]
 Genome-wide association studies have been hugely successful in identifying dozens of common genetic risk factors for a large number of common diseases. However, one area that GWAS has not had much success in is the field of psychiatric illness, where finding common risk factors that replicate across studies has been consistently difficult. However, it looks like this is starting to change. The current issue of Nature Genetics has two papers from the Psychiatric GWAS Consortium, detailing some of the largest meta-analyses of schizophrenia and bipolar disease ever published.
Genome-wide association studies have been hugely successful in identifying dozens of common genetic risk factors for a large number of common diseases. However, one area that GWAS has not had much success in is the field of psychiatric illness, where finding common risk factors that replicate across studies has been consistently difficult. However, it looks like this is starting to change. The current issue of Nature Genetics has two papers from the Psychiatric GWAS Consortium, detailing some of the largest meta-analyses of schizophrenia and bipolar disease ever published.
The schizophrenia study robustly replicated two previously implicated variants, and discovered five new ones, and the bipolar disease study replicated one and discovered a new one. The new variants give us some pretty startling insights into the genetics of the diseases, in particular revealing the importance of a non-coding gene micro-RNA 137 in regulating a wide range of genes expressed in neurons. As always, these variants explain only a small proportion of the total genetic effect, but they show that psychiatric genetics has now truly entered the GWAS arena, with all the scientific benefits that this can bring to medical research. [LJ]
The images above, in order, are taken from the paper Temporal Trends in Results Availability from Genome-Wide Association Studies, and from Wikimedia Commons.
 In this week’s Nature Genetics there is a genome-wide association study of lung disease severity in cystic fibrosis suffers (or at least the subset who carry the ΔF508 mutation). The authors report a number of variants with “suggestive evidence”, and one with genome-wide significant evidence . The one genome-wide significant variant is
In this week’s Nature Genetics there is a genome-wide association study of lung disease severity in cystic fibrosis suffers (or at least the subset who carry the ΔF508 mutation). The authors report a number of variants with “suggestive evidence”, and one with genome-wide significant evidence . The one genome-wide significant variant is 



 RSS
RSS Twitter
Twitter
Recent Comments