 On Monday, the Guardian published an article by plant geneticist Jonathan Latham entitled “The failure of the genome”. Ironically given this is an article criticising allegedly exaggerated claims made about the power of the human genome, Latham does not spare us his own hyperbole:
On Monday, the Guardian published an article by plant geneticist Jonathan Latham entitled “The failure of the genome”. Ironically given this is an article criticising allegedly exaggerated claims made about the power of the human genome, Latham does not spare us his own hyperbole:
Among all the genetic findings for common illnesses, such as heart disease, cancer and mental illnesses, only a handful are of genuine significance for human health. Faulty genes rarely cause, or even mildly predispose us, to disease, and as a consequence the science of human genetics is in deep crisis.
[…] The failure to find meaningful inherited genetic predispositions is likely to become the most profound crisis that science has faced. [emphasis added]
The claim that human genetics is in crisis is not novel. Latham made an extended version of this argument in a blog post at the Bioscience Resource Project in December last year, which Daniel critiqued at length at the time, and which contained a schoolboy statistical error corrected by Luke. And Latham is by no means the only genome-basher out there: the 10 year anniversary of the sequencing of the human genome triggered a spate of “genome fail” pieces (see Nicholas Wade, Andrew Pollack, Matt Ridley, and a particularly horrendous example from Oliver James, for instance).
We suspect for most of our readers Latham’s rather hysterical critique will fall on deaf ears, but it is part of a bizarre and disturbing trend that needs to be publicly countered. Here are several of the places where Latham’s screed gets it patently wrong:
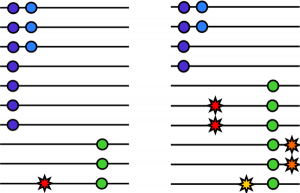
 Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe.
Last week, a post went up on the Bioscience Resource Project blog entited The Great DNA Data Deficit. This is another in a long string of “Death of GWAS” posts that have appeared around the last year. The authors claim that because GWAS has failed to identify many “major disease genes”, i.e. high frequency variants with large effect on disease, it was therefore not worthwhile; this is all old stuff, that I have discussed elsewhere (see also my “Standard GWAS Disclaimer” below). In this case, the authors argue that the genetic contribution to complex disease has been massively overestimated, and in fact genetics does not play as large a part in disease as we believe. I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ]
I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ] RSS
RSS Twitter
Twitter
Recent Comments