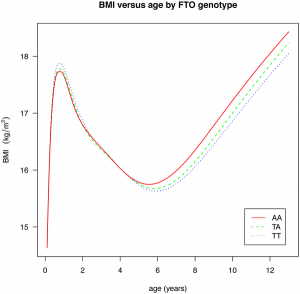
 There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
It’s budget battle season in the United States, and biomedical research funding looks likely to be caught in the crossfire. President Obama has proposed a $745 x 106 increase in the NIH budget, bringing it to $31.8 x 109 in total. This wouldn’t quite keep up with inflation, leading to a slight decrease in grant success rates from America’s largest biomedical research funder. The Republican-controlled House of Representatives, by contrast, has slashed the NIH budget by $1.6 x 109 in their proposed budget (bill HR1), which would be a heavy blow to research funding. Of course, scientists, non-crazy editorial writers and activist groups have been rallying around protecting research funding (in the NIH and beyond). Unfortunately I wouldn’t expect a speedy resolution, as veteran US politics blogger Nate Silver likens the whole situation to Zugzwang. [JCB]
 I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ]
I have to say, this isn’t going to be an easy job; assembling a high-quality reference genome of an under-studied organism is a lot of work, especially using Illumina’s short read technology, and identifying and making sense of tumour mutations is equally difficult. Add to this the fact that the tumour genome is from a different individual to the healthy individual, this all adds up to a project of unprecedented scope. On the other hand, the key to saving a species from extinction could rest on this sticky bioinformatics problem, and if anyone is in the position to deal with it, it’s the Cancer Genome Project. [LJ] RSS
RSS Twitter
Twitter
Recent Comments