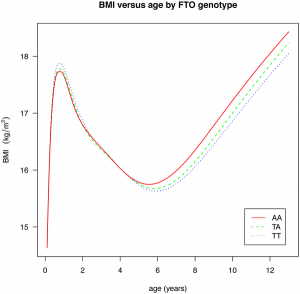
 There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
It’s budget battle season in the United States, and biomedical research funding looks likely to be caught in the crossfire. President Obama has proposed a $745 x 106 increase in the NIH budget, bringing it to $31.8 x 109 in total. This wouldn’t quite keep up with inflation, leading to a slight decrease in grant success rates from America’s largest biomedical research funder. The Republican-controlled House of Representatives, by contrast, has slashed the NIH budget by $1.6 x 109 in their proposed budget (bill HR1), which would be a heavy blow to research funding. Of course, scientists, non-crazy editorial writers and activist groups have been rallying around protecting research funding (in the NIH and beyond). Unfortunately I wouldn’t expect a speedy resolution, as veteran US politics blogger Nate Silver likens the whole situation to Zugzwang. [JCB]
Manuel Corpas (creator of the personal genome visualisation tool myKaryoView) has a handy post reviewing various tools for exploring personal genetic data, including SNPtips, Enlis Genome, and his own browser. He concludes (fairly) that none of these tools provide everything needed for investigating your own genome; there is still plenty of room for clever coders to produce a killer app in the personal genome space. [DM]
There is an interesting qualitative study in the European Journal of Human Genetics this week looking at how parents with genetic disease go about explaining the nature of genetic risk to their children. The authors interviewed 33 families with a range of genetic diseases, including Huntington’s disease and cystic fibrosis. There are some pretty heart-rending conclusions. Parents will often delay talking to the child about the disease until they are older, but children themselves tend to be at least partially aware from a much earlier age. Openness and honesty was a great help to the children, but parental anxieties from various sources can make this difficult. Explaining inheritance patterns to children is difficult, even for dominant diseases, partly down to the difficulty of convaying probability, though the use of pie charts seems to help somewhat. A good example of this is how often “a 50% change of getting the disease” translated into “either me or my sibling will get the disease”. [LJ]








 RSS
RSS Twitter
Twitter
0 Responses to “Predicting lupus outcomes, US biomedical funding battles and telling children about genetic disease”