 I have no strong family history of any disease, despite having 7 blood aunts and uncles and countless cousins. So when I sent my spit off to 23andMe at the start of the Genomes Unzipped project, I was expecting something very similar to Caroline’s experience: a 5% increase in risk here, a 2% decrease in risk there, nothing that would really tell my anything about my health.
I have no strong family history of any disease, despite having 7 blood aunts and uncles and countless cousins. So when I sent my spit off to 23andMe at the start of the Genomes Unzipped project, I was expecting something very similar to Caroline’s experience: a 5% increase in risk here, a 2% decrease in risk there, nothing that would really tell my anything about my health.
However, this was not my experience. Along with a pretty interesting Y haplogroup, I also had three unexpected and potentially worrying health results. I am a cystic fibrosis carrier, a hemochromatosis compound heterozygote, and have a strongly elevated risk of age-related macular degeneration. This cocktail of genetic disease certainly was not what I came to the test expecting!
After some thinking, I decided to take my test results to my GP, and see if there was any advice or testing he would recommend. In the end, my GP referred me to a clinical geneticist, which started a cascade of appointments which in turn led to a number of important changes in how I treat my own health.
What was most interesting is how the whole experience got me thinking about my health as something I am in charge of. I have since made a number of important life-style changes, some of them directly related to my genotyping results, some more generally to improve my overall health.
The point of this post is just to go through some of the experiences, what I have learned about specific conditions, and what changes I have made to my life since. In some sense, I feel like my experience is a case-study in what good outcomes can come from personal genomics, both for specific conditions, and more generally for how genetic data can change your own approach to your health.
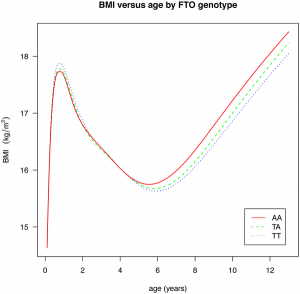 There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ]
There are a pair of papers in PLoS Genetics that shine some light on the effect of common GWAS variants on complex traits. The first investigates the effect of 22 common variants on sub-phenotypes of systemic lupus erythematosus, in how well a model including clinical measures plus GWAS variants can predict specific complications of lupus, over a model including just clinical outcomes. In some cases, there is little improvement (GWAS variants add nothing to prediction of renal failure, for instance), but in many there is a measurable improvement (such as for hameatological disorder and oral ulcers, the former of which is largely unpredictable from clinical outcomes). The second paper is a breakdown of the effect of the common obesity-associated variant FTO on BMI across age ranges; we see an interesting effect, whereby the variant that causes an increase in BMI in older children actually causes a fall in BMI in children under the age of 2. [LJ] A couple of interesting articles this week on the Personal Genome Project and public genomics in general. Mark Henderson at the Times has an opinion piece (behind a paywall, I’m afraid) about Misha Angrist‘s book Here Is A Human Being (see also this review from The Intersection), and in the Duke Magazine Mary Carmichael has an in-depth feature on the work of George Church, with some interesting history of the early days of the PGP.
A couple of interesting articles this week on the Personal Genome Project and public genomics in general. Mark Henderson at the Times has an opinion piece (behind a paywall, I’m afraid) about Misha Angrist‘s book Here Is A Human Being (see also this review from The Intersection), and in the Duke Magazine Mary Carmichael has an in-depth feature on the work of George Church, with some interesting history of the early days of the PGP. Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ]
Two exciting-looking new science blogging collectives have been announced this week. The Public Library of Science launched a new blogging collective, including personal genomics blogger Misha Angrist, and the Guardian newspaper has launched its Guardian Science Blogs network, including Dr Evan Harris, ex-MP for Oxford West and long time supporter of the role of science in public policy. I’m pretty excited about these new blogs, but it does stand to increase my RSS load significantly. [LJ] Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say:
Finally, Procreation News; our very own Daniel MacArthur and Ilana Fisher have recently given birth to a baby boy (the picture to the left may be a little out of date). Daniel made the announcement on Twitter, and also had this to say: RSS
RSS Twitter
Twitter
Recent Comments