 Just out in prepublication at PNAS is a paper from Eric Lander’s lab, entitled, somewhat provocatively The mystery of missing heritability: Genetic interactions create phantom heritability. The authors suggest that certain types of gene-gene interactions could be causing us to underestimate how much of the heritability of complex traits has been uncovered by our genetic studies to date.
Just out in prepublication at PNAS is a paper from Eric Lander’s lab, entitled, somewhat provocatively The mystery of missing heritability: Genetic interactions create phantom heritability. The authors suggest that certain types of gene-gene interactions could be causing us to underestimate how much of the heritability of complex traits has been uncovered by our genetic studies to date.
There has been an awful lot of talk about this research since Eric Lander talked about it at ASHG a few years ago, and the paper itself has generated quite a bit of discussion on- and off-line. Razib Khan reported on the paper last week, giving a good summary. He mentioned a press release about the paper issued by the advocacy organisation GeneWatch, which confuses the additive heritability discussed in this paper with the total heritability of diseases (a distinction explained below), and uses this to draw conclusions about how this result alters the promise of personal genomics. This just goes to show how much confusion there already is out there about this subject.
I have a more detailed post up on Genetic Inference about this paper, the strength of the argument, and what it means for the field. Here I am just going to pull out what I think are some important take-home points about this paper:
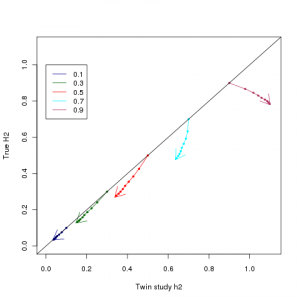
1) Broad sense heritabilities (the kind that are clinically important for e.g. risk prediction) have NOT been significantly overestimated The type of heritability we ultimately care about, the broad or total heritability, is how much total phenotypic variation is captured by genetics, or equivalently the correlation between identical twins in uncorrelated environments. The figure at the top of this post shows a plot that I made using Zuk et al’s equations, comparing true broad sense heritabilities, against what would be estimated based on twin studies (I have matched the colouring etc to Figure 1 of the paper). The twin study estimator of heritability is a robust estimator of total heritability for heritabilities less than 0.5. Above that, LP epistasis causes growing overestimation – it can make a 50% heritable trait look like a 65%, and 70% look like a 95%. It does not make weakly heritable traits look strongly heritable, just strongly heritable traits look very strongly heritable.
2) This paper is discussing additive heritability. This is a specific form of heritability that acts “simply” – half of it is passed on to offspring, siblings share an amount proportion to how related they are, and the genes that underlie it do not interact with each other. We do not know how much heritability acts like this, but various lines of evidence have made us think that it is a relatively good model, and most competing models have been incompatible with this evidence, or look contrived. What Zuk et al have done is produce a set of plausible, simple and non-contrived models (Limiting Pathway or LP models) that look pretty much indistinguishable from additivity using many of the tests we have run, but can act very differently in twin studies. Under these models, twin studies will overestimate the additive heritability (i.e. make us think that a larger proportion of heritability acts “simply”). The equivalent plot to the top of the page for estimating additive heritability, which you can see here, shows massive overestimation of additive heritability across the spectrum.
3) There is no real evidence that these LP models apply (and in fact there are still a few reasons to believe additivity could still broadly apply, see my other post for details). The issue is that we cannot conclusively rule these models (or models like these) out, and therefore the heritability explained by the genetic variants we have found so far is very uncertain.
4) This is important because our measures of “heritability explained” by the genetic variants we have found look at how much additive heritability is explained. These measures have in general told us that we have only explained a small proportion (generally < 25%) of additive heritability – but if in fact the heritability is largely not additive, but we are treating it like it is, we could in fact have explained a higher proportion of heritability than we believe. This would mean that the “missing heritability” is missing not because we have not found the right genetic risk factors, but because we have not found the right model to use. This could be good news: the genetic variants we have discovered could in fact be used to predict disease a lot better than they we can at the moment, if only we can find the right model to use them with.








 RSS
RSS Twitter
Twitter
There’s an interesting paper (for me) that high recombination rates exclude epistatic effects: http://www.pnas.org/content/106/16/6866
“Here, we present a computational model of selection dynamics involving many epistatic loci in a recombining population. We demonstrate that a large number of polymorphic interacting loci can, despite frequent recombination, exhibit cooperative behavior that locks alleles into favorable genotypes leading to a population consisting of a set of competing clones. When the recombination rate exceeds a certain critical value that depends on the strength of epistasis, this “genotype selection” regime disappears in an abrupt transition, giving way to “allele selection”—the regime where different loci are only weakly correlated as expected in sexually reproducing populations.”