Gholson Lyon is a physician-scientist currently working at the Utah Foundation for Biomedical Research and the Center for Applied Genomics at Children’s Hospital of Philadelphia. He will be starting as an assistant professor in human genetics at Cold Spring Harbor Laboratory next month. I asked him to write this guest post to provide some personal context to his thought-provoking commentary in Nature (subscription required) on returning genetic findings to research subjects. [DM]
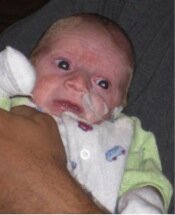
Photo of Max, who died aged four months from Ogden syndrome. Posted with permission from his family.
I have just published in Nature a commentary discussing the need to bring exome and genome sequencing into the clinical arena, so that these data are generated with the same rigorous clinical standards as for any other clinical test. This way, we can then easily return at least medically actionable results to research participants. In this day and age of consumer and patient empowerment, I can also see eventually returning all data, including the raw data, to any interested participants, as this can then promote crowd-sourcing for data analysis, with research participants controlling and promoting the relative privacy of and analysis of their own data.
As I described in my commentary, my thinking on this matter was prompted mainly by Max (see picture) and his family. The obituary for Max can be found here, and that of his cousin, Sutter, here. We described their condition here, and we named this new disease Ogden Syndrome in honor of where the first family lives. I am now trying to think about and discuss the human aspects of and lessons from this story. My thinking has also been influenced somewhat by the late James Neel, who wrote a very thought-provoking book called Physician to the Gene Pool.
To me, it was deeply disconcerting that I could not officially return any results to this family (or to another family in a different project discussed here) even when the papers describing the genetic basis of their disease were published, as this was considered “research” and was not performed in a clinically appropriate (CLIA-certified) manner. This was all the more painful when one of the sisters in the Ogden family became pregnant and asked me what I knew. I cannot predict whether it would have helped or hurt this woman to learn during her pregnancy that she was indeed a carrier of the mutation, with the associated 50% risk of her baby boy having the disease. I also do not know if she would have undergone any genetic testing via amniocentesis of the fetus prior to birth (with the associated ~1% risk of miscarriage from the procedure), nor do I know what decisions she might have made prior to the birth even if she had undergone such testing. All in all, it was certainly an ethical and moral dilemma for me not to be able to return the research result to her, given that the results were not obtained in a CLIA-certified manner. It is still an issue, as there are even now financial and systematic barriers for getting all women in the family tested with a CLIA-certified gene test for NAA10 (which was developed over a six month period by ARUP Laboratories). It would have been so much better if we had just done the entire sequencing up front in a CLIA-certified manner.
It is therefore my opinion that the current flood of human sequencing data is not being optimally generated, given that there is no regulation requiring the initial sequencing of each human to be performed in a clinical-grade manner, thus making it very difficult and unwise for anyone to return any research information to research participants. This is due to the fact that clinicians must “first, do no harm”, so returning less than clinical grade sequencing results to patients can potentially cause such harm, as noted above. Therefore, the natural corollary to this argument (to me at least) is that initial sequencing of human exomes and genomes should be performed in a CLIA-certified (or other clinical-grade) manner up front, so that the data can be returned to research participants and/or linked to their medical record. That way, as knowledge expands over the next few decades, we can constantly go back and re-analyze the genomes and update research participants on new unrelated findings, in the context of their biochemical individuality. Of course, one can always re-sequence using newer technologies and/or confirm any important results on a new DNA sample from each person, but there will no longer have to be rigorous development of a specific CLIA-certified gene test for each mutation, given that the exomes and genomes were sequenced up-front in a clinically proper manner.
What are the benefits?
The medical system in America is currently geared and financially rewarded to treat specific issues that people have when they are ill, whereas there are relatively few incentives in the system for counseling (genetic and otherwise) and keeping people healthy. This needs to change. A federal guideline mandating that the first exomes and genomes on each human be performed in a CLIA-certified or otherwise clinically appropriate environment could save substantial money in the long run, for many reasons, in my opinion, including in the area of preventive medicine. This of course includes beefing up the number of genetic counselors initially, but there is accumulating evidence that at least some people take corrective actions (some of which can be life-saving) when they learn of genetic predispositions. Also, preimplantation diagnosis and other preventive efforts may benefit readily from the increased carrier screening that will accompany the clinical sequencing of humans.
Problems with the current system
Some researchers have suggested that the “easy” thing would be to “simply” re-sequence the research samples for each mutation of importance in an already CLIA-certified lab. However, this overlooks many things, including 1) not obtaining the blood/saliva or isolating, storing and tracking the DNA in any sort of specified and reproducible manner, 2) not developing an official CLIA-certified Sanger-sequencing test for this particular gene, and 3) not interpreting the Sanger sequencing in any sort of CLIA-certified manner. It is not enough to just place a sequencing machine in a “CLIA-certified lab” and then simply declare that all DNA, no matter how that DNA was obtained, stored, or tracked, nor how the sequencing data were analyzed, will suddenly now yield “CLIA-certified” results, deliverable back to research participants. What about the real risks of sample mix-up and incorrect results? In addition, are researchers really going to take the time to develop CLIA-certified tests for each and every one of the possible medically-actionable mutations that they uncover now?
Conclusion
Even though it is very likely that whole genome sequencing will become a routine part of clinical care in 10-20 years, it is far from certain what the path and process toward this goal will be. In science, much as in public policy, there is sometimes Brownian motion, whereby there are many small steps forward and backward, with many people losing perspective on the overall progress being made. Thirty years ago, very few people would have predicted that in vitro fertilization and test tube babies would become a routine part of medicine, even garnering a Nobel Prize for one of the champions of this technology. Furthermore, targeted carrier screening and pre-implantation genetic diagnosis is also becoming a routine part of medical care, particularly in some communities with an excess burden of recessive genetic diseases. So, the real question is: where we will be in two years? And in five years? The current sequencing will certainly lead to many scientific papers and discoveries, but the question is whether any of this sequencing, genetic discoveries, or unrelated findings will actually directly benefit anytime soon the actual families who took the time to donate their blood and other tissue samples? Will results, including unrelated findings, be returned to these families so that they can take preventive measures? Will the genomic data be returned directly to research participants so that they control the privacy of and access to their own genomic data? If so, how will this be done to minimize error and misinformation?
Hopefully, one day soon, each person will possess a copy of their own clinically certified genome, and they will then have the option to link their genome to their medical record and/or to provide their own genome data to other researchers. Indeed, some people could even contribute to the analysis as citizen scientists, akin to what is described in the new book Reinventing Discovery: The New Era of Networked Science by Michael Nielsen. Many researchers and interested participants can then together analyze the genome of each person repeatedly, as our collective knowledge expands. The consent process could also follow the model being proposed by the Sage Bionetworks Common Genomic Research Project, including a version of “portable legal consent“. Right now is hopefully a revolutionary time in medicine, as discussed by Eric Topol in his new book The Creative Destruction of Medicine, so we should seriously consider doing the right thing now, before we have gone down this current path far too long.
Afterword
I am struck by the following quote in a recent Nature editorial:
What kind of work deemed as accepted today will be denounced by future generations? The question is one that all researchers should bear in mind, because history may judge them more harshly than their peers do.
Currently, it is the wild West for human genome sequencing. Many researchers are sequencing as many exomes and genomes as possible, using a range of methods. However, I am struck by the question of whether we are really thinking through the ethics of this. In the 1940’s, there was a huge rush to find a cure for syphilis and other venereal diseases, and this led many prominent researchers to conduct experiments on humans that we now look back on as being morally repugnant. Are we repeating the same mistake when we sequence live humans in research environments in which we are not able (or perhaps willing) to inform and counsel these people concerning potentially very deleterious mutations in their genomes? Are we keeping these people “in the dark” and thereby taking away from them the chance to take preventive measures for highly penetrant mutations running in their families, i.e. their “clan genomics”?
Acknowledgements: First and foremost, I would like to thank the families with whom I have had the pleasure to work. The Ogden Syndrome family also graciously consented to the publication of the photograph.
Conflicts of Interest: I do not have any obvious conflicts of interest to declare. I have had informal discussions with representatives from Illumina, Sage Bionetworks, ARUP, Golden Helix, Complete Genomics and Omicia, Inc., but I have not had any formal consulting role, nor have I received financial compensation from these or any other companies performing DNA collection or sequencing.








 RSS
RSS Twitter
Twitter
The title of your Nature piece is:
Whereas the title of this piece is:
These are markedly different. It is possible to agree with the second, but not the first.
People doing clinical-grade genetics are kinda busy. Do they want to waste their time on the worried well(ish) when there are desperately serious cases like the ones you refer to, to be diagnosed?
But where are the findings that clinical genetics relies on going to come from, if research-grade research stops?
We have to assume that the price of doing clinical-grade research is much, much higher than doing research-grade research: is the subsequent loss of research a price worth paying? For example, I would anticipate that none of the samples used in any of the WTCCC projects meet the tracking and storage requirements that can absolutely guarantee results come from the correct subject. We do our best, but know that is not the same thing. Are we therefore to stop large-scale genetics research, for fear we find something that places us in a moral dilemma? Note – that is not a rhetorical question, but genuinely an option facing some research groups.
Perhaps we should ask the research subjects what they want? i.e. are you taking part in this research in order to get a diagnosis? If so let’s do clinical grade research – or refer you somewhere that does it. If not, are you prepared to take part in genetic studies knowing that we will not feed back information to you; or will only feed back information if we know it to be life-threatening, and with the caveat that you will need to be re-tested by something more clinical grade?
That is quite sad that people so hang up on the concept of “research” grade finding vs “CLIA-certified” findings. If you are not sure in the quality of your genetic research so why bother publishing it? On other side if you really found something disturbing like the case described in the paper it is researchers moral and ethical obligation to consult with genetic counselors and facilitate independent validation in another laboratory even it if is not CLIA-certified. Could could have saved a lot of pain for these families by taking a bit more personal responsibility and being more proactive. I understand the reluctance in the view of legal issues, but every University and Hospital has an able legal team that could be consulted with in a due time.
Some people forget that the primary goal of research is to help people and not publish paper and wait when someone else will pick the tab to implement research findings.
I feel that I should respond to the above comments, although I will be addressing such issues in future talks and commentaries as well.
I would argue that I was MUCH more proactive than most researchers in this context, because I actually met the family, got the CLIA-certified test developed, am working to get them tested and counseled, etc…. There are many issues involved, which I have tried to begin addressing in the commentary and this posting, mainly to get more discussion going on this subject. Also, please just read the Retraction Watch website to discover how much “research” is of poor-quality, for many reasons. Also, I don’t remotely agree with you about “saved a lot of pain for these families by taking a bit more personal responsibility”, as I would challenge you or anyone else to indicate to me what more I could have done in this extremely challenging situation? I am trying to help researchers to become more aware of these issues, because you can avoid all this by doing the sequencing up front in a CLIA-certified or clinical-grade manner, or alternatively you could be VERY explicit in your consent documents about NON-return of ANY results and/or complete de-identification of samples (making this completely 100% research only), but of course losing out on the ability to go back to these families for future information. I say this latter thing because it is a two-way street, because if one goes back to families or medical records for updated information, this means you then should take responsibility for conveying important clinical-grade findings back to these families.
I agree that one alternative is that the informed consent documents could be very explicit regarding non-return of ANY research results, including explicit example statements such as “If we find a Huntington’s disease mutation or highly penetrant breast cancer mutation, we will NOT tell you about this, as this is research only and you will derive ZERO benefits from this research”, and there should be no grey zone in between, lest we mislead participants into thinking that we actually plan to help or counsel them in any way about their genetic findings. Or, the samples can be completely deidentifed with ZERO intent for re-identification, which should also be made very clear to anyone asked to participate in such a study. However, if there is the chance for re-identification by the researchers, then return of results must be discussed explicitly and very concretely with participants, in terms of return or NON-return of anything. Otherwise, one enters into this slippery slope of how to return results reliably and appropriately if the sequencing was not done up front in a clinical-grade manner.
I have inserted below some typical language that I found in one IRB-approved informed consent protocol for collecting genomic DNA for research, so that I can deconstruct the implications of this language with the reader.
Example language:
1) “It is possible that we will uncover disturbing information about you or your family during this research. For example, information about paternity or adoption might be discovered. We will not reveal this information unless it has direct medical significance for your family.”
2) “BENEFITS. We hope that this study will help us better understand the condition that affects you, however there may not be any direct benefit to you. If we find anything of medical significance to you or your family during this study, we will inform you in writing and provide you with an opportunity to speak with a medical geneticist or genetic counselor to explain the results.”
3) “NEW INFORMATION. If we learn something that might affect whether you want to be in the study, we will tell you. We plan to report the results of this study to you or your family. We will try to keep you informed about progress of the research.”
Such language in an informed consent is very well-intentioned, but also very misleading to the participant, as it is not clear to me that this has been well-thought out in terms of how research results can be returned in any sort of meaningful or systematic way, given that they are not obtained in any sort of clinically regulated manner, i.e. CLIA-certified in America. Researchers might face major risks of lawsuits if they return incorrect or misleading results to research participants, plus giving research-grade results back to people is basically practicing bad medicine without a license, which is why I am asking that the sequencing be performed in such a manner as to involve clinicians. We have agencies and policies like FDA, Medicare, and CLIA to ensure that test results and other things offered to people meet certain high standards; otherwise, there is substantial risk of misinformation and misguided “counseling”.
In addition, are researchers really going to take the time to develop CLIA-certified tests for each and every one of the possible medically-actionable mutations that they uncover now? [/quote]
Actually, why not have standards, the data brought to the clinic has to be free from these ambiguities that we have come to tolerate in sequencing what with tweaking the data and what with increasing the coverage or read depth, a clinical diagnosis is not like a ROC curve where there are grey areas, it has to be a clear cut diagnosed versus non-diagnosed with enough evidence to tell the patient and deliver an impact on their treatment options.
In sequencing, the problems start from the platform, go on to the analysis, culminate in the assembly of the genomes afterwards and some of these errors are inherent in the system and others are batch-driven so clinicians can’t really tolerate that fuzziness and of course it is so justifiable.
Back to what I quote from your article, ‘TIME’, is it ‘time’ or ‘willingness’, or maybe a combination of both that may influence the demotivation that you felt? Many of the breakthroughs were made because an issue affected the inventor directly (probably his offspring suffered a genetic disability) and they were keen into getting to the bottom of it, e.g is the IonTorrent story. So I highly doubt that ‘time’ is the matter here, bioinformaiticians and folks in the NGS arena are probably too much burrowed into the joy they experience in their own field rather than seeing areas of applications such as the clinical field for them to shape-up, I am a bioinformatician myself however,I will side with the notion that it comes from humans – the data that we are fascinated with- and we have to bring it back to them in the form of a credible assessment of their genetic makeup.
Clinicians like Stephen Kingsmore are engaged into developing a pipleine for finding disease causing variants and they wanna standardize the analysis process in a form befitting the clinical requirements.
Which sequencing? All sequencing?
The next post on this site goes into great detail into how healthy people harbour major loss-of-function genetic variants. If someone has, or is suspected to have, a rare genetic disorder, these are just the things we would look for and want to feed back. Without that motivation, there is no reason to think these variants are majorly harmful to the individual concerned. The first scenario needs a clinician and genetic counselling, the latter doesn’t.
I agree that none of the major sequencing projects currently have anything in place to adequately track the samples and data to any particular individual, at least not to the high standards required of clinical tests. This is exactly the problem. As I alluded to above and in my commentary, I am asking that the INITIAL exome and whole genome sequencing of each human should be performed in a clinical-grade manner, so that at least medically actionable results can be returned to families, in which their “clan genomics” are known, i.e. there is evidence that these mutations are causative for disease in the context of their genomes and environment. Ideally, I would like to have all clinical-grade genomic data returned to participants so that they can decide whether to link the data to their medical record for future analysis and/or to release their data to other researchers, thus empowering and engaging the participants in a meaningful way. Researchers can then sequence and re-sequence the DNA as much as they like in a research setting, with explicit consent from participants that no such information will be returned, but I am asking researchers and policy-makers to consider that it should be necessary and required to perform at least clinical-grade sequencing up front for the first exomes or genomes on any live humans participating in human genetics research, so that results can be easily returned with appropriate counseling. In America, return of results requires CLIA certification; in other places it would require approval from other bodies; and in some settings (e.g. some developing countries) where the regulatory environment is unclear, it would simply require improving the standard of sample collection testing to reduce the incidence of sample swaps and other false results.
At the moment, the only place to my knowledge in America offering CLIA-certified whole genome sequencing is at Illumina, but Complete Genomics is applying now for CLIA-certification for their whole genome sequencing, plus several companies, including 23andMe, are now offering CLIA-certified exome sequencing. To the best of my knowledge, in the case of Illumina, the process involves drawing the blood in a barcoded tube sent to the ordering physician by Illumina, followed by shipping the blood to Illumina for CLIA-certified DNA extraction and sequencing, which dramatically minimizes any possibility for sample mix-up. Their bioinformatics pipeline also originates from the CLIA-certified environment, and their list of variant calls is necessarily stringent to lower the number of false positives. From a research and discovery perspective, this might mean missing some possible variants due to certain regions of the genome not having adequate coverage for a reliable call, but the raw data are available and can always be re-analyzed in a research setting and/or with improving CLIA-certified bioinformatics pipelines. There is a certain economy of scale and reliability (think Amazon) with centralizing CLIA-certified whole genome sequencing with companies such as Illumina and Complete Genomics, but I suspect that other companies and institutions will nonetheless want to offer this service themselves too. So, it is simply a matter of establishing minimal guidelines that must be met in order for such places to generate CLIA-certified exomes and whole genomes. The bottom line is that CLIA-certified sequencing is already available now, and it is time to embrace this in a clinical setting, so that the fruits of the human genome project can finally start to help families and patients on a broader level.
If I understand correctly, ogden syndrome is a recessive x-linked disease for which the authors knew that the maternal grandmother of the male fetus was a carrier. If the mother knows that she is a carrier also, her probability of giving birth to a sick boy is 50%. On the other hand, if the mother does not have this information her subjective probability is 25% and if the mother knows for sure that she is not a carrier the probability approaches zero.
The expert choose to withhold information from the mother leaving her with the subjective probability of 25% of having a sick baby. The question now becomes this: what was the true level of knowledge of the expert (as opposed to the mother)? Lets suppose that the probability of the test showing a mutation in the right position when this position is in fact wild-type is 5% (a fairly sloppy research scientist might be assumed to mislabel or otherwise mix up 1 test tube in 20, sequencing error-rate must be much lower). Lets further suppose that the probability of the test outcome being “wild-type” when the mother is in fact a mutation carrier is also 5%.
Using the Bayes rule we can calculate that the expert should now believe with 95% certainty that the mother is a carrier.
If the expert believed that only 1% error-rate applies, then the level of his belief should rise to 99%.
The mother may know very little about genetics but this is surely not a valid reason to deprive her of the chance of interpreting the statement: „it is over 95% probable that your baby boy will have a fifty-fyfty chance of having the disease“? Or put differently: would it be a good idea to wait several months in order to be able instead to say: „it is over 99,9% probable that your baby boy will have a 50:50 chance of being sick“? It seems doubtful to me that the difference between 95% and 99,9% would be enough to change the mother‘s behaviour.
I’m all for barcoding samples, but does it really benefit research subjects to use a licensed phlebotomist? To withhold diagnoses that are high-confidence, although not certain, because no gene diagnostic test is licensed? To withhold a diagnosis for months because the subject does not have insurance coverage for genetic counseling? It all seems incredibly paternalistic.
I’m also confused about reagent tracking. Assuming that most of the relevant data is from second-generation sequencing, how will reagant tracking prevent errors in diagnosis? It doesn’t sound very plausible — is there something I’m missing?
I just spent the past three days attending this molecular diagnostics conference, attended by many specialists in the world of gene diagnostic testing http://www.triconference.com/. One thing I confirmed for myself is that there is extreme variability in the turnaround time for various “CLIA-certified” gene tests, and some organizations claim to be able to develop such tests quickly and effectively in a matter of weeks. This was certainly not the case for me with the very reputable diagnostic laboratory that I approached, as it took 6 months to develop and make available a single Sanger-sequencing based gene test for NAA10, for MANY reasons, including financial reasons in terms of getting them to commit the resources necessary to develop the test and officially offer it with sign-off by board-certified medical geneticists.
In terms of the above reader comment, medicine has always been very paternalistic, although one purpose of my commentary is to empower patients and consumers by making it possible for them to obtain a clinical-grade genome, at their request. Currently, any patient is allowed to request a copy of their medical record, including laboratory test results. I have done this myself, so I have copies of results from my electrolytes, cell counts and liver function tests. Likewise, if someone sequenced my genome, I would like to be able to receive a copy of my genome, including raw data, on a small terabyte hard drive, so that I can share it with whomever I choose for further analysis. I am a participant in the Personal Genome Project, based out of Boston, so I hope to receive back my genome, if and when they sequence me. But, we have rules and regulations to maintain decent standards AND to impede quacks and other unethical types “peddling” shoddy or fraudulent products. I am therefore calling for proper clinical standards to be applied to the sequencing of the first exome and/genome from each live human being, so that so that such exomes and genomes really can be the same as a “lab test” returned to the patient or research participant. Right now, in the current situation, most genomes being sequenced do NOT meet the criteria in place for laboratory tests.
To reinforce the above point, given the fact that some research (see Retraction Watch website) is either not reproducible, poorly done, ill-conceived or even outright fraudulent, the critical current issue relating to the “return of results” is to ensure that any exome or genome sequencing of human beings is performed in an appropriate CLIA-certified (or equivalent) clinical environment, with rigorous standards in place, including for sample collection. It is only then that clinicians will be able to return results in any sort of meaningful quantity to research participants, given that it is currently incredibly tedious and bureaucratic to develop one-by-one CLIA-certified gene-based tests to validate mutations.
One cliffhanger is that our health care system, at least in America, is so badly broken that 8 months have gone by and the doctors for one of my research subjects, in which I discovered the unrelated finding explaining his idiopathic hemolytic anemia (see my Discovery Medicine paper), have still not ordered a CLIA-certified test for the mutations in PKLR causing this person’s anemia. Apparently, this gene test is officially available only in Germany and the Netherlands, as per the doctor’s office, and they don’t have the time to track down any company in America willing to develop the test. It will likely also be difficult (as per usual) to get approval from insurance to reimburse the cost of the test. So, he still doesn’t know that he is a carrier for these mutations, even though he is likely going to get married and have kids soon. To me, it is absolutely crazy that our health care system is unable and/or unwilling to invest enough money and time in genetics testing, counseling and prevention. This is one of many reasons that I am requesting that the first exome and whole genome sequencing in each human should be completed in a CLIA-certified clinical environment, so that researchers can return such information to participants and so they don’t have to face or go through what I am still dealing with. Plus, I imagine many lawsuits will occur once research participants start passing on mutations that were uncovered in research studies but not relayed back to the research participants. Some researchers just tell me that I should give back my research results to these participants, but that is BREAKING THE LAW, given that all clinical genetic testing in America is regulated under CLIA, and my results were NOT generated in a CLIA environment. I am trying to do the right thing here, including publicizing these major problems with the way in which current exomes and genomes are being sequenced, at least in America.
“Some researchers just tell me that I should give back my research results to these participants, but that is BREAKING THE LAW” — I sympathize. I hope your story will push the FDA to reform some of these rules.
Just FYI, this was also covered over the weekend here:
http://www.sltrib.com/sltrib/news/53503970-78/research-family-lyon-born.html.csp?page=1
I am probably not as up-to-date as I used to be when writing a blog on genetic testing, but–at the time–CLIA certification was for quality of laboratories and testing and had no specific genetic testing standards. For example, there was no requirement that a “gene” for condition X be proven to cause condition X and certainly nothing to say that a specific DNA sequence be matched to a specific condition. Many genetic testing laboratories available over the Internet may advertise their CLIA-certification, but that doesn’t guarantee that their genetic testing is accurate or correct.
To address the comment from Marie, the point is to have minimal clinical standards in place for the first germline genome from each human, so that the genomic data can be returned to the participants. There is simply a need in human genetics to re-establish the researcher-participant contract, which will certainly be enabled by the return of results. An easy way to return results has already been demonstrated by 23andMe, with their very useful interface, which could easily be adopted for the delivery of whole genome data back to participants, as long as the sample collection and sequencing is performed in a CLIA-certified manner in America. This is a much more distributive model, whereby research participants really do participate in the analysis of their genomes, and it relieves researchers of the burden of having to “return results all at once”, as it is much easier to analyze and re-analyze the clinical genomes, as knowledge expands. The bottom line is that clinical whole genome sequencing is coming very quickly, so I anticipate that this will sort itself out as more people realize just how important it is to return results to participants, particularly those results of high medical impact within particular family structures. This is in the same vein of thought as the concept of “Clan Genomics” articulated in Cell. 2011 Sep 30;147(1):32-43. Clan genomics and the complex architecture of human disease. Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA.
Just as an aside, if one does a Pubmed search for “clan genomics”, the only other paper of relevance is this below paper, which is also very interesting:
Proc Natl Acad Sci U S A. 2010 Jan 26;107 Suppl 1:1779-86. Epub 2009 Sep 23. Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Bittles AH, Black ML.
I am just cross-posting my response to a Letter to the Editor that appeared in Nature yesterday. I posted this same thing to the Nature website, but I want to put it here as well. Here you go:
However, I would just point out here that I am only asking for the INITIAL germline genome for each human to be sequenced with appropriate clinical standards, so that each human who is sequenced can have their genome given back to them and hopefully linked to their medical record, with their consent. Whole Genome Sequencing is a disruptive technology that can finally help bring forth individualized medicine, as discussed in Eric Topol’s new book, The Creative Destruction of Medicine, but this may only come from consumers, rather than the biomedical research complex.
The sequencing and return of genomic data can go a long way toward implementing some carrier screening for highly penetrant mutations, some of which can have relative risks of 7,500-10,000x (acknowledgement here to following reference: Genet Med. 2012 Feb 16. doi: 10.1038/gim.2011.78. [Epub ahead of print] Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Biesecker LG.)).
The bottom line is that potentially many humans will carry such mutations, and it would be enormously useful to catalog human genetic variation by actually sequencing and returning whole genomes to humans in a clinical setting. A further benefit could be to centralize these genomes in one database, so that it is MUCH easier to calculate penetrance of disease with certain mutations. As I say, this is a disruptive technology, particularly if done to scale in centralized facilities that are specialized for whole genome sequencing, and the time is fast approaching when this can and should be done in at least developed countries.
Also, the Retraction Watch website documents what I believe is just the “tip of the iceberg” in terms of research that is outright wrong and therefore a complete waste of money, and I would argue that this is because there are no set or required standards for good high-quality research. Requiring that all initial germline genomes be performed at the level of Good Clinical Practice seems like a great idea to me, so that we can at least get high-quality genomes going forward. Right now, many sequencing centers are sequencing thousands of germline exomes and genomes, but not with any clinical standards in place, so all of that data cannot be returned to research participants, plus the raw sequencing data are not being freely shared, at least not readily. To me, that is a huge waste of money, particularly given that people who take the time to donate their blood and saliva typically want to hear about their own results.
Of course, we can argue about when exactly will be a good time to sequence everyone’s genome in a clinical setting, but I sure hope that in 20-50 years time, every infant born (at least in industrialized countries) will have their genome sequenced, so that preventive and therapeutic services can finally be offered on a more individual level. I am just trying to do my little bit to move that time closer to 20 years, in contrast to 50 years! I would recommend people read Eric Topol’s book, along with Michael Nielsen’s book on Reinventing Discovery: The New Era of Networked Science. I have attached some other references in my posting at Genomes Unzipped.
Since the publication of this commentary, I have had MANY people tell me that this is indeed a major problem that needs to be addressed, given the therapeutic misconception that many people have when they enroll in a human genetics study, no matter what they are told. I am attempting to head off a huge crisis that might occur when and if many research subjects are not informed about highly penetrant mutations discovered in their exomes and genomes by researchers. I would highly recommend people read the story of one patient and now patient advocate Rebecca Fisher, as per below:
A closer look revisited: are we subjects or are we donors? Rebecca Fisher, MLIS, Genetics in Medicine, advance online publication 23 February 2012. doi:10.1038/gim.2012.6.
Truth and information should be put way above politics and allow
a person to make their decisions based on KNOWLEDGE. It’s not fair
for the researchers to play god or withhold information they have
discovered from a potential parent who may have to deal w/ the possibility of passing this gene forward. Be honest, screw politics
I think what’s missing from this discussion is the need for clinical validation of genetic markers.