In May of last year, Li and colleagues reported that they had observed over 10,000 sequence mismatches between messenger RNA (mRNA) and DNA from the same individuals (RDD sites, for RNA-DNA differences) [1]. This week, Science has published three technical comments on this article (one that I wrote with Yoav Gilad and Jonathan Pritchard; one by Wei Lin, Robert Piskol, Meng How Tan, and Billy Li; and one by Claudia Kleinman and Jacek Majewski). We conclude that at least ~90% of the Li et al. RDD sites are technical artifacts [2,3,4]. A copy of the comment I was involved in is available here, and Li et al. have responded to these critiques [5].
In this post, I’m going to describe how we came to the conclusion that nearly all of the RDD sites are technical artifacts. For a full discussion, please read the comments themselves.
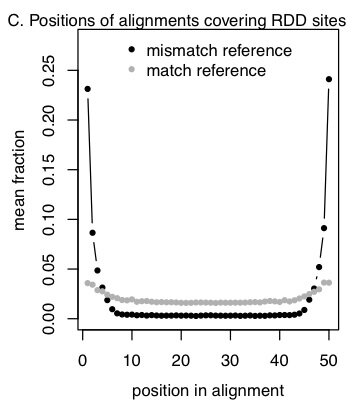
Position biases in alignments around RDD sites. For each RDD site with at least five reads mismatching the genome, we calculated the fraction of reads with the mismatch (or the match) at each position in the alignment of the RNA-seq read to the genome (on the + DNA strand). Plotted is the average of this fraction across all sites, separately for the alignments which match and mismatch the genome.
Continue reading ‘Questioning the evidence for non-canonical RNA editing in humans’
 RSS
RSS Twitter
Twitter
Recent Comments