From genetic association to genetic test
We’ll start with defining our genetic test. For the sake of argument, imagine we’ve done a genome-wide association study of a sample of patients with a serious polygenic medical condition called Madeupitis (thanks to Luke for drawing my attention to this important disorder), and a sample of people without the disorder. We’ve found a set of genetic variants associated with Madeupitis, and we’d like to use them as the basis of a predictive genetic test. First we need to characterize the test’s ability to classify people into two groups: those who are at high risk of Madeupitis, and those who are not. We compare our test’s classification results to a diagnostic “gold standard”, in this case, diagnosis by a clinician specialising in Madeupitis. This comparison gives us the sensitivity and specificity of our test.
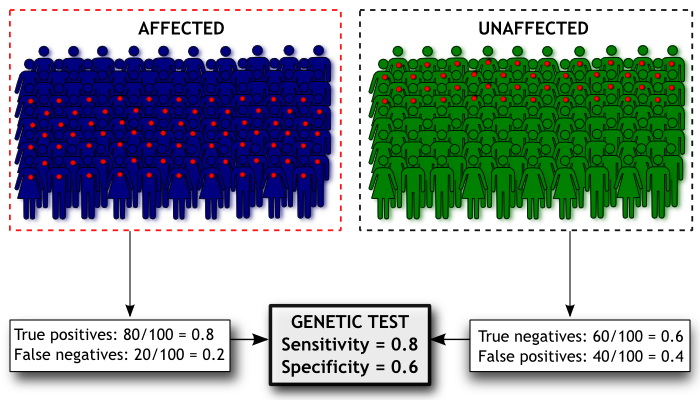
Sensitivity concerns our study participants who have Madeupitis. It is the proportion of people with the disorder that our test correctly classifies as having Madeupitis. This is the proportion of “true positive” test results. Specificity concerns the people who do not have Madeupitis. It is the proportion of people without the disorder who are correctly classified by our test as not having Madeupitis. This is the proportion of “true negative” results; 1-specificity therefore gives us the proportion of “false positive” results for the test (the proportion of people without Madeupitis who had a positive test result). If sensitivity and specificity both have a value of 1, the test correctly classifies everyone; in practice all tests have some degree of error, but we would still like these values to be as close to 1 as possible.
Here’s a diagram of how we’d estimate the sensitivity and specificity of our Madeupitis test. In our study we had 100 people with Madeupitis (coloured dark blue) and 100 healthy people (coloured green). When we administered our genetic test, 80 people with Madeupitis had a positive genetic test result (indicated by the red dot), but 40 of the healthy people also had a positive genetic test result. This gives us sensitivity of 0.8 and specificity of 0.6 for our test.
From genetic test to population screening
Sensitivity and specificity are helpful for giving us an idea of the discriminatory power of our test, but they don’t actually tell us about its validity as a screening test in the general population (i.e. not our research sample). To get an idea of this, we actually want to look at the problem from the opposite perspective. Before we were interested in the probability of someone having a particular genotype given that they had (or didn’t have) Madeupitis; now we want to know the probability of someone developing Madeupitis given that they have a particular genotype. This sounds like semantics but they are different things. Here’s an example…
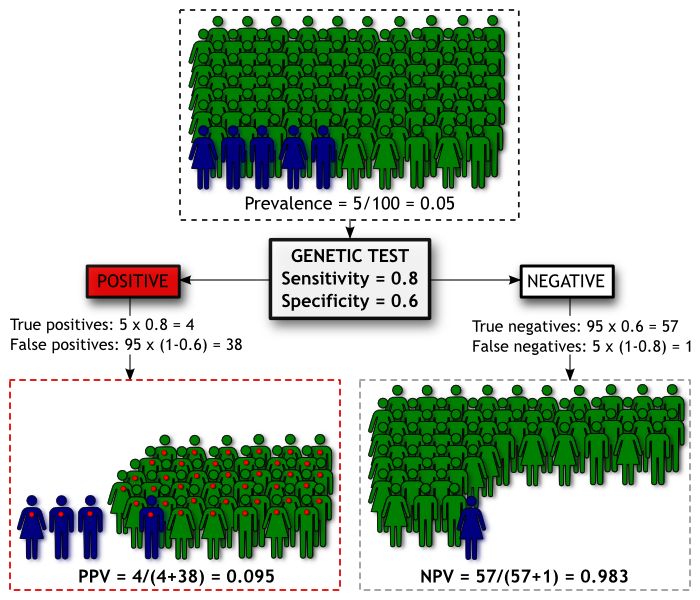
We have a population of 100 people. We know from epidemiological studies that the prevalence (proportion of people with a condition) of Madeupitis in this population is 5%. This means 5 people will develop Madeupitis and the other 95 will not. Our genetic test has moderate discriminatory power, with sensitivity of 0.80 and specificity of 0.60. We can summarise the screening results in the diagram below, where people coloured blue are those who will develop Madeupitis, and those coloured green will not. As before, a red dot indicates a positive genetic test result.
Using our test we will identify 42 people as being likely to develop Madeupitis. Of these people, 4 will be true positives and the remaining 38 will be false positives. The Positive Predictive Value (PPV) is the proportion of people with a positive test result who go on to develop the condition, which is 0.095 for our test in this population. Our test will also categorise 58 people as being unlikely to develop Madeupitis, 1 of which will be a false negative. The Negative Predictive Value (NPV) is the proportion of people with a negative test result who don’t develop Madeupitis, which is 0.983 in this population.
Unfortunately this means our test isn’t doing a good job of identifying cases. If you’re given a positive result, we’re not very confident about whether you’ll get Madeupitis or not (our PPV of 0.095). Put another way, for a member of this population, the pre-test probability of developing Madeupitis is 0.05 and the post-test probability (for those with a positive test result) is ~0.10 – not a great improvement in prediction. On the other hand, if you’re given a negative result, we’re pretty sure you’re not going to get Madeupitis (our NPV of 0.983). Whether the NPV or PPV of a test is more important really depends upon the consequences of a negative or positive result for that test. But let’s say that in this instance we’re concerned that those 38 out of a 100 people who are given false positive results might go out and unnecessarily spend a lot of money on expensive treatments to protect themselves against Madeupitis. How can we improve the performance of the test?
Improving PPV and NPV
The easiest way to improve PPV is to increase the prevalence of Madeupitis in the population. Obviously we can’t do that directly, but we can avoid testing people who have a low likelihood of testing positive, based on some other source of information. Let’s say we only offer our test to people who have at least one parent who has Madeupitis (knowing that Madeupitis is influenced by genetics, this seems a plausible criterion).
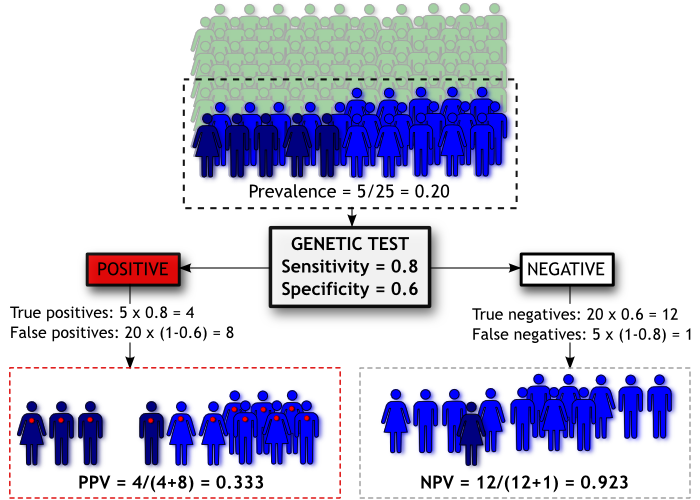
Going back to our diagram, we have now “pre-screened” our population based on family history. The people coloured green have no family history and are not administered the genetic test. People coloured light and dark blue all have a family history of Madeupitis and are therefore given the test, but as before only those coloured dark blue will go on to develop the disorder.
By filtering our population on family history we effectively increase the prevalence of Madeupitis in the subset of the population we’re testing (from 5% to 20%), improving our chances of correctly classifying people. Thus our PPV increases to 0.33 even though the sensitivity and specificity of our genetic test haven’t changed. However, there is a trade-off; our NPV has been reduced to 0.923. This is called “cascade testing” and it is sometimes used to determine whether people should have a genetic test for a particular disorder (e.g. Huntington’s disease).
The other thing we could do is improve the sensitivity and/or specificity of the genetic test itself. We might do this by including additional variants in the test, or we might combine our genetic test results with information on other non-genetic risk factors. For example, if we want to estimate risk of cardiovascular disease, we might find we have more predictive power if we combine genetic risk factors with known environmental risk factors such as smoking.
What does this mean for personal genomics?
First of all, it doesn’t mean that DTC genetic tests aren’t useful. At the moment the sensitivity and specificity of a lot of genetic tests for complex, polygenic disorders (for which we haven’t yet identified all the genetic variants that increase risk) are unlikely to match those of standard diagnostic or screening tests. What’s likely is that the predictive capacity of these tests will improve as more variants are identified, and/or if additional non-genetic information is included in the test. The important thing is to keep concepts such as PPV and NPV in mind when you look at DTC test results and remember that for complex diseases, the results you get are always probabilistic not deterministic.
The diagrams in this post were made using Inkscape, an excellent free software package. You can re-use the text and/or diagrams from this post as long as you acknowledge that they’re from Genomes Unzipped (under a Creative Commons Attribution-NonCommercial 2.5 Generic License).









 RSS
RSS Twitter
Twitter
Great post, Kate.
I guess an important distinction also needs to be made between the utility of a test from a public health perspective (clinical utility) compare to an individual perspective (personal utility). Virtually nothing currently offered by personal genomics companies would meet the requirements for being offered by (say) the NHS, but it can still be useful for individuals interested in deciding where to invest their effort in terms of health monitoring and prevention.
It would be interesting to get a discussion going some time about the best way to think about personal utility; a good topic for a future post!
Excellent.
For a real-world example of someone attempting to use a DTC result, Blaine Bettinger posted about this earlier this year:
http://www.thegeneticgenealogist.com/2010/01/07/personalized-genomics-a-very-personal-post/
Yes personal utility is an important issue whereas the focus has usually been on clinical utility. Specific and narrowly defined clinical utility is probably not really offered by whole genome scanning (except for BRCA and a few others), but personal utility goes much further. Just reading about your results and how genetic variants interact with environment to influence health is already a personal benefit I would argue. Maybe you are more likely to read even generic looking health information when your genetic results are included in the reports, this should be useful. Personal benefit can also be sparking an interest in a new subject, satisfying curiosity, feeling that you are learning something. Almost certainly personal benefits cannot be “proven” through randomised clinical trials!
So I agree – it would be a good idea to have a more full discussion about it, it’s a wide area, is touched upon here and there but I have not yet found any detailed articles about it. Genomes Unzipped is becoming an extremely useful resource for background information and would be the perfect place.
BTW – forgot to add, very useful post and thanks for the link to Inkscape!
This traditional medical genetics approach is appropriate for diseases and other traits that are objectively defined and discrete — the disease is either present or absent and the cutoff between the two states is essentially non-arbitrary. Rare recessive diseases like cystic fibrosis mostly fall into this realm.
But most complex traits are not discrete. Cholesterol levels, blood sugar, blood pressure, height, weight, and so on, are quantitative traits, and the line between healthy and unhealthy is relatively arbitrary.
The potential utility of personal genomics for a disease like Type II diabetes is not to provide a “positive” or “negative” reading, but to provide risk assessment. In an ideal world, a genome scan would provide the likelihood (a value between zero and one) that an individual with a particular genome will develop a more or less severe version of the disease, at a particular age and in a particular environment. This is a very different way of thinking about human genotype and phenotype.
@Lee
I think you’re confusing two concepts here. One is that certain diseases don’t have sharply defined borders (such a T2D), the other is that genetic risk for complex disease does not diagnose a disease, merely gives a probably that someone will develop the disease (the ideal world you speak of is our world, and a probably between 0 and 1 is exactly what a genetic risk prediction gives you).
Either way, neither of these concepts are particularly new. Epidemiologists have been investigating risk factors for decades, and identifying which groups of people have elevated risk of getting the disease, and even using population level family data to quantify the probability of an individual developing the disease conditional on a relative having it.
Anyway, regardless of how fuzzy your definitions or your probabilities are, from a personal or public health perspective at some point you have to choose to make an intervention for high-risk individuals (scheduling regulation mammograms, proscribing blood thinners, etc), and this will usually require you to draw a line in the sand. As soon as you do this, questions need to be asked about about how effective the test, including the cut-off you apply, are at predicting whatever outcome you are trying to avoid (metastatic cancer, heart failure, etc). Kate’s post is still very relevant.
Thanks everyone for some nice discussion and interesting points.
As Luke says, the usefulness of any of these statistics depends upon what you want to use a test for. The overall objectives of a public health screening programme and a DTC genetic testing service are different. But if individuals take DTC tests that give them an assessment of disease risk, one of the possible outcomes is that they’ll decide to seek medical advice or treatment for those conditions they appear to be at high risk of developing (although the critical level of risk that prompts a visit to the doctor may differ between people).
The way we assess public health screening tests isn’t a perfect framework for evaluating DTC genetic tests, but it’s a place to start. I agree with Daniel that those of us who are interested in this field need to think about how to assess the personal utility of these tests. Definitely something we’ll be discussing more in future posts.
@Luke
I don’t disagree with you on the science. My concern is with the potential for traditional medical genetics terminology to cause confusion — among members of the public — when applied to complex personal genome results.
First, the word “diagnosis” has been used traditionally at the level of phenotype not genotype. If we ignore fuzzy disease borders and variable expressivity, I agree with you that a diagnostic result is either “positive” (person has disease) or “negative.” And as Katherine explained, we can evaluate the accuracy of a specific “diagnostic test” by counting true positives and false positives, true negatives and false negatives. The same logic holds true for traditional genetic tests directed at Mendelian conditions defined by a 1:1 correspondence between genotype and phenotype.
Problems arise, however, when this traditional terminology is used to describe interpretations of complex, late-onset disease risk based on personal genome profiles. What exactly is being diagnosed genetically in a pre-symptomatic individual? Is it the risk of future disease (speculative phenotype) or the presence of complex genotypes that increase the risk? The GAO guy certainly didn’t know, but neither does anyone else outside the biomedical sphere.
If you’re diagnosing genotypes, then yes, you can use a simple positive/negative scale. Furthermore, as we know, the accuracy of genotype results from legitimate companies (e.g. 23andMe) is extremely high.
But if your “diagnosis” refers to a potential future phenotype with incomplete penetrance, the use of “positive” and negative” to describe genetic results makes no sense (for the public). It’s this misunderstanding that is responsible for headlines like “Personalized gene tests are bogus? Of course they are” on an opinion piece written by the bioethicist Art Caplan for MSNBC.
IMHO, if we want rational regulation over DTC genomics, we have to have policy makers who understand the difference between genotype and phenotype, the meaning of penetrance, and the difference between simple Mendelian inheritance and complex inheritance.
Luke — what’s your take?
@Lee,
Very good points. Not all sickle cell patients have crises.
But all have sickle cell mutation. Not all CF patients are infertile.
But all have CFTR mutations. Not all heart attack patients have 9p21.3 SNP changes,
not all 9p21.3 have heart attacks. This is incredibly tricky. Which is why it may be silly to not regulate according to
“Is it related to disease and health?”
Otherwise, we may be arguing over grains of sand and get nothing done. I understand ardent proponents of DTCG will want is to argue this minutiae but I think it is a folly.
The question is this “medically related or not”
@Lee
Yeah, I agree that the idea of diagnosing a complex disease using genetics is a nonsense, because your genes only define a probabilistic risk of developing the disease. However, my point is that there is nothing new about this; genetics is just another source of disease risk factors.
No-one thinks that being a smoker is a diagnosis for lung cancer, but it is definitely a strong risk factor, and is thus used as a basis for public health intervention. Likewise, if you are over 50, do less than 30 minutes of physical activity a day, and have a waist circumference greater than 40 inches, you should be taking actions to avoid or mitigate against T2D, even if no-one is going to use these facts to diagnose you.
Basically, we have a perfectly good public health framework for considering risk factors. I don’t share your worry about potential confusions, at least not more than I do with the “Smoking Kills” labels on cigarettes.
Steve,
You keep making the same trivial argument over and over again, without ever actually explaining exactly what rules we should be using to delineate between medical and non-medical tests. This isn’t contributing anything useful to the discussion. Move on, or I’m going to start deleting your comments.
With regards the optimistic-ish conclusion that:
I’d refer you to David Clayton’s paper last year:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2703795/?tool=pubmed
Prediction and Interaction in Complex Disease Genetics: Experience in Type 1 Diabetes.
PLoS Genet. 2009 July; 5(7): e1000540.
which argues that the PPV would remain low for most complex diseases, even if all relevant loci are found.
Very nice post and overall a great blog! Just a few comments on the last paragraph:
From the perspective of clinical utility, Katherine Morely’s argument “we might combine our genetic test results with information on other non-genetic risk factors” could be turned around into the concept that we can use family or personal health history to identify individuals who may benefit from predictive genetic testing.
Familial adenomatous polyposis (FAP) is an example: individuals with a mutation in the APC gene have an almost 100% lifetime risk of developing colorectal cancer compared to a lifetime risk of 1/20 in the general population. FAP is rare, colon cancer not so rare, and knowing your carrier status for the APC gene mutation can result in a life-saving preventive removal of the entire colon and rectum. So fishing out that rare individual who will benefit from genetic testing based on family history can have dramatic implications.
Conversely, looking at complex disorders such as type 2 diabetes mellitus (T2DM), considering the small effects of disease risks variants does not yet provide clinically useful information in the sense that it would change the management (an individual with a family history of T2DM on maternal and paternal side already has an increased risk of developing DM of 4.0 compared to baseline population risk).
In addition, combining the relatively small effects of disease risk variants with substantially larger effects of phenotypic features for risk prediction in complex diseases is not so straightforward and previous attempts to combine the genotype score with clinical models (such as in the Framingham Heart study) did not result in a significant improvement of the predictive power. And although I agree with Katherine that “the predictive capacity of these tests will improve as more variants are identified” other factors may be important and we need to learn much more about the pathophysiologic roles of the identified risk variants as well as their interaction with genetic and environmental processes. The phenotype as a composite model may already incorporate the effect from known or yet undiscovered risk alleles. Environmental factors, such as obesity and diet, might have a much stronger influence on the development of t2Dm than genetic factors.
Family history is often used as an argument against clinical utility of personal genetics, claiming that genetics offer no extra predictive value. I don’t think it’s completely valid, one thing is that different generations often live in completely different environments. It may be true that the predictive value of genetics is currently low, but it is NOT zero and can be improved if other factors are incorporated (see bit.ly/bB2Efd and bit.ly/d0n4jq). Also genetics and family history are complementary rather than completely overlapping – in the case of Celiac Disease for example if there is family history the advice is to take a (not very predictive) genetic test to see whether or not the individual has inherited those “family” genes. See also the case of Blaine Bettinger and diabetes (bit.ly/4Chszs) where the genetics appeared to confirm his FH risk.
Yes the tests can be entertaining and fun, but they are not totally useless from a healthcare point of view. Not very predictive is still better than not predictive – small differences and small lifestyle changes make a big difference over the 10, 20, 30 years of complex disease development.
Getting to this a bit late – sorry!
I think the question of how strongly predictive tests for complex diseases might be if/when we manage to identify the majority of the relevant loci is an interesting one. Naomi Wray and colleagues present some data (PLoS Genetics 2010) that suggest more optimistic conclusions than David Clayton, showing that for some complex diseases such as type-1 diabetes and Crohn’s disease it might be possible to develop a test using all relevant loci with an AUC of >0.98, and thus a decent PPV. But as Clayton points out, these theoretical predictions are partially dependent upon the estimates of lambda_s used, which may not always be particularly accurate. Theoretical projections aside, I think the predictive power of these tests will improve to some extent as we identify more loci, and that treating genetics as one of a set of risk factors used to identify those at high risk might improve predictive power further for some diseases.
That said, I’m skeptical about whether a test for complex disease predisposition based purely upon genetic information will ever achieve a high enough PPV to be implemented as a population-wide public health screening test (but I’ll happily eat my words if that happens). Additionally, the results from some efforts to combine other known risk factors with currently identified genetic risk factors have only shown a modest improvement in predictive capacity (e.g. Lango and colleagues for type-2 diabetes, Wacholder and colleagues for breast cancer).
Even if predictive tests for complex diseases don’t achieve the PPV needed for public health screening, the information they provide may be useful for the individual in a medical context, at least for some diseases. But (not to labour the point) we need to think about how to evaluate that…
I agree that family history and genetics (as well as personal health and environmental history) should be used in a complementary fashion. As it is true for personalized medicine, many commonly used risk assessment tools are only based on epidemiological data without being validated as predictors of disease, so their clinical utility remains uncertain. Instead of the debate regarding personal genetics and family history, efforts should be focused on how to harmonize both research strategies.
For a number of disorders including the familial cancer syndromes, a good family history represents the first genetic screen followed by appropriate genetic testing. In other instances such as a type 2 diabetes mellitus (T2DM), the magnitude of risk changes derived from family history as well as personal and environmental history (obesity is the single most important risk predictor for T2DM) is so much greater that – at least at this time – the predictive values of genetic risk variants are clinically not very useful. So I think every condition has to be evaluated individually regarding the best available approach to meaningful risk prediction.
As mentioned by Keith, Celiac disease (CD) is a good example where simple clinical models using easily accessible parameters such as family history are not sufficient for risk assessment and of the roughly 1% affected individuals in the general population, many go undetected potentially causing significant morbidity. Although highly accurate serologic tests are available for diagnosis of CD, they may not be diagnostic in the pre-clinical stage. Here, the information from several chromosomal celiac disease risk regions identified in recent GWAS may indeed be used clinically to improve risk prediction in the future.
I agree with Keith’s last point. I think the significance of genetic risk variants derived from genome-wide approaches reaches far beyond risk assessments and for contributes to our pathophysiologic understanding of complex diseases. To stay with the example of diabetes, the genes corresponding to GWAS-derived risk variants cluster in several distinct metabolic pathways which may lead to new pharmacogenetic approaches to drug development.
@Daniel,
Delete comments?
Daniel, the statement is clear. Regulate Medical as Medical. Delete away if it makes you feel good. I will tehn just post the same thing on my blog and say you deleted it under the guise of useless commentary, when in fact I say you are too chicken to address it.
No? I think the whole field is too chicken to address it.
No?
Ok, then tell me why no one has addressed this issue?
Why can’t anyone answer this question. Why can’t anyone except me address it?
I argue it because NO ONE answers it. Yet this is precisely what the field will come to dude.
What do you define medical testing as?
Maybe we should start there. We can ask lawyers, PhDs and VCs what they think medical tetsing is. We can collect the answers and then do the same for physicians, counselors, regulators and insurers.
How hard is it to get you guys to stop blathering about the horror of regulation or the pain of getting the SNPs cut off from a marginal business model now?
Address the question. What do you think medical testing is?
I for one am dying to know.
And no, I am not going to jade you with my answer first.
Regulate medical as medical. End of story. The judge is clearly the FDA and the government. If you don’t get out there and start saying what you think medical is, these companies will have a very narrow range of testing.
Haircolor and earwax and asparagus sniffer will be what’s left…..
Oh and athletic prowess ;)
Get it?
-Steve
Nevermind Daniel,
Don’t respond, just read my blog post. Then you will get it…..
http://thegenesherpa.blogspot.com/2010/08/what-is-medical-testing-why-it-matters.html
Regulate medical as medical. End of story. The judge is clearly the FDA and the government.
No. The FDA is prohibited by law from regulating the practice of medicine. It only regulates medical devices and drugs. It does not regulate physicians, surgeries, supplements, medical records…
Why can’t anyone answer this question.
Because the FDA itself has no idea what a medical claim is. Take it away, Alberto Gutierrez
That’s the MONEY quote at the end. Usually the FDA gets away with vague generalities like that. Oh, the “rules are worked out pretty well”? Ok, regulate away! Most people are taken in by that head fake and stop there.
But unusually for a reporter, there is a slight detour into specifics, at which point it is revealed that the guy in charge of enforcing the policy “doesn’t know where the policy is” and admits it “won’t be that easy for people to follow it”!
Steve
You have a very particular debating technique. The fact is that this what is medicine question HAS been addressed by many people, including myself, Daniel, Dan and many other fowls – in many blogs including your own. But that’s your style, you say things for effect. You say everything is down to you. You say a million things and then when one of them comes true a few years later you say “I told ya so … if only you had listened to me”
Your style is to say that many are “blathering about the horror of regulation” while the truth is that most of us are not against regulations but rather are trying to debate what type of regulation or oversight is best for all. But you know that very well because you read my blog, Daniels, Dans and this one.
Anyway, it’s not “what is medical testing” but what is a medical device, as Steven points out (and it looks as if the FDA have already decided on that – without answering the question).
I point all this out hear because GenomesUnzipped is a new forum and it is certainly attracting a new readership who may be unfamiliar with the territory.
Medicine matters, healthcare matters, that’s why it’s reasonable to discuss risks and benefits before over regulating a particular sector.
@Keith,
It is now clear, DTCG is a device.
LDT is going to be regulated.
The only say that DTCG will have is to define what a medical claim is.
LDT on the other hand will have no ability to do this. Why? They HAVE always been medical.
My point is, the debate of what is medical matters because it is at the root of medical claims, medical devices and medical testing.
I keep saying regulate medical as medical because that is what should be done.
I don’t think any of you have done a good job saying what that is.
That’s all.
As for Oprah’s genome scan….called it ;)
@Steve Yarrow. Mary puts the links provided in the article. One is a list of definitions for the cosmetic act. Which lays out what a drug or device is. The other is basically a Boolean look up for all FDA policy……
I agree, the FDA cannot regulate medicine. That is the territory of the states and now the federal government.
The problem most people have with this whole defining Medicine issue is that most people have no freaking clue what medical doctors do. Most either watch it on TV or read about it. PHDs “rotate” on some wards, unless they do clinical work.
Regulators? Maybe if they are practicing physicians.
In most legal definitions I have come across there is always some gray zone of what medicine is.
This gray zone effectively allows it to be argued that most things dealing with health, healing, maintaining well being, disease, palliation, human suffering, and changes in “normal” physiology are medical.
Hence, what is medical? Just about everything deing with the above mentioned can be medical.
This is the debate. And we all need to have it. Otherwise, someone else will.
Steve,
[Comment cross-posted to Gene Sherpas.]
In case readers here are unaware of the context of this threat: you’ve posed the same question in no fewer than seven separate comments on Genomes Unzipped, and it’s no less trivial now that it was when you first asked it. Forgive us if we’re getting a little sick of it.
This is not the crucial question here; indeed, it’s nothing more than semantics. We shouldn’t be regulating on the basis of whether or not a test fulfils some arbitrary definition of medicine (put forward, I add, by someone with a financial stake in maintaining the status quo).
Instead, we should consider regulation on the basis of more pragmatic questions:
1. Are the results of this test conveyed accurately to consumers?
2. Are false claims made by the providers of the test for marketing purposes?
3. Are customers provided with sufficient information to assess the validity and utility of the test results?
4. Is there evidence that this test, as currently provided, will do harm to recipients?
5. Is there evidence that this test, as currently provided, will benefit recipients?
These are the questions that matter, Steve – and your focus on this irrelevant semantic issue is nothing more than a distraction from the interesting discussion that needs to be had: how can we best ensure that testing companies provide accurate results, refrain from false advertising, and provide transparent, accessible information about test validity and utility? How do we gather the information required to assess the benefits and harms experienced by test recipients? And how do we achieve these goals without stifling innovation or inhibiting the public right to self-education and exploration of their own genome?
@Daniel
Your pragmatic questions are absolutely on-target.
As long as the first four are answered Yes, No, Yes, No for a DTC product, we don’t have to worry about the fifth because it will be decided by the market. If people continue to pay to receive personal information, then those people obviously consider it a benefit. It doesn’t matter what anyone else thinks.
hi!!!