 Illumina CEO Jay Flatley announced that an upgrade to their HiSeq 2000 platform expected this spring will allow users to generate 600 gigabases of sequence (the equivalent of 5 high quality human genomes) per one-week run of the machine. This would essentially double the current throughput of the platform and propel Illumina even further ahead in the arms race of delivering vast quantities of low cost sequence data. [JCB]
Illumina CEO Jay Flatley announced that an upgrade to their HiSeq 2000 platform expected this spring will allow users to generate 600 gigabases of sequence (the equivalent of 5 high quality human genomes) per one-week run of the machine. This would essentially double the current throughput of the platform and propel Illumina even further ahead in the arms race of delivering vast quantities of low cost sequence data. [JCB]
Over at Golden Helix, Gabe Rudy has just completed a three-part series introducing readers to the promise and challenges of new DNA sequencing technologies, which is well worth a read for those just starting out in the analysis of next-gen sequence data or who have a more-than-casual interest in the current state of the field. [DM]
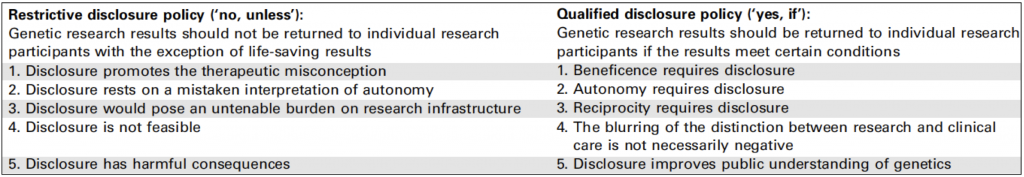
This month’s edition of Trends in Genetics includes a review article on the ethical issues raised by the feedback of individual genetic data to research participants by Bredenoord and colleagues. This has long been a subject of debate, but the recent increase in studies that assay a large number of genetic variants (such as genome-wide association studies and whole-genome sequencing studies) has brought this issue to the fore. There is currently no consensus on how to deal with this, and in my experience the approach favoured has varied both between projects and between the ethics committees that have assessed them.
Bredenoord and colleagues take the view that the issue is not if results should be disclosed but what type of results should be disclosed and how this should be achieved. From their review of the literature they see two schools of thought on this: restrictive disclosure and qualified disclosure (see the table from their article below).
The “what” and the “how” of individual feedback are issues of debate in themselves, and could change the way we do research (e.g. by increasing the involvement of clinicians and genetic counsellors in research studies). Regardless of whether you agree with the authors’ perspective, the article makes an interesting jumping-off point for further debate. [KIM]
What’s in store for the future of personalized medicine regulation? In a guest post at Xconomy, Dan Vorhaus explains why a recent New York legislative proposal to mandate insurance coverage of genetic tests for cancer is symptomatic of larger problems within the U.S. regulatory environment which, according to a recent report, may be responsible for a shifting global balance in medical technology innovation. [DV]








 RSS
RSS Twitter
Twitter
I think the TiGs article is the wrong frame and sets up a false dichotomy. I guess I know what my guest post for DG Mac will be about…
@Misha
I really only have a surface understanding of the debate regarding return of individual data, but I tend to agree; the two viewpoints didn’t really seem to be in direct opposition to me. Look forward to reading your views on the topic!