 When Daniel first asked me if I wanted to be involved in Genomes Unzipped, I was one of the more hesitant participants. I weighed up the pros and cons, but in the end what sold me was that after almost a decade of curiosity I finally had the opportunity to find out my genotype for the hereditary haemochromatosis (HH) variants in the gene HFE. But things didn’t unfold quite how I’d expected, and I’m still left with some unanswered questions about HH in my family.
When Daniel first asked me if I wanted to be involved in Genomes Unzipped, I was one of the more hesitant participants. I weighed up the pros and cons, but in the end what sold me was that after almost a decade of curiosity I finally had the opportunity to find out my genotype for the hereditary haemochromatosis (HH) variants in the gene HFE. But things didn’t unfold quite how I’d expected, and I’m still left with some unanswered questions about HH in my family.
A little bit of background to HH
HH causes your body to store too much iron, potentially causing organ damage if not detected promptly. Unlike many common diseases, HH is not influenced by a multitude of genes. The most common form of HH is associated with variants in the HFE gene that change amino acids in the protein it encodes. The variant with the most severe effect on the protein is C282Y, although two other variants, H63D and S65C, also appear to affect its function. Disease severity varies between people carrying the same HFE variant, however, suggesting that there may be some as-yet unidentified genetic factors that modify the effects of these major mutations. Mutations in HFE are most commonly found in populations of Northern and Western European descent, although the variants exist in populations from Southern Europe, Asia, and Africa as well. 23andMe only report results for the C282Y and H63D variants in relation to HH.
HFE and my family
My father is of Anglo-Celtic ancestry and has a family history of HH, so when he was found to have an extremely high level of iron in his blood, the diagnosis was obvious. Genetic testing determined that he carries one H63D mutation in HFE. I was pretty certain that my 23andMe test would show me to be a H63D mutation carrier as well, and not just because of family history. I had a health check-up before leaving Australia for the UK that showed my ferritin levels (a measure of the amount of iron in the blood) were now above the normal range for women.
Prior to taking the 23andMe test I talked to my family about what it might tell us. They were unworried and supportive, and a little curious about whether Dad had passed his H63D mutation on to me. But my 23andMe test revealed something unexpected: I discovered that I have not one, but two copies of the H63D mutation. Which means that my mum also has at least one H63D mutation, something I subsequently explained to my family via a Skype call to Australia.
I did wonder whether my test result was correct; the genotyping standards for DTC genetic tests are very high, but there is always some chance of error. But then my sister had a clinical genetic test for all three major HFE variants and found that she also has two copies of the H63D mutation. My sister also being homozygous confirmed that our mum is a carrier.
Maybe it’s not just about HFE
Any readers who know a bit about HH will by now think there’s something weird about my family. Only about 1% of people who are homozygous for the H63D mutation will go on to develop iron overload. It’s even more unlikely for someone like my dad who has only one copy, but when he was diagnosed his ferritin levels were more than five times those of the average man. It’s also strange that I have high ferritin levels, given that I’m a healthy woman in my 30s. On the other hand, my mum and my sister have ferritin levels well within the normal range.
As I mentioned above, iron overload severity varies between individuals with the same HFE genotype and it’s been hypothesised that this could to be due to the presence of other genetic factors that modify the effect of the HFE variants (although environmental factors obviously play a role as well). This seemed a plausible explanation to me for what’s going on in my family; maybe my dad and I carry some other genetic variants that increase our risk of iron overload.
Researchers only started intensively investigating this hypothesis in the last decade so there isn’t a lot of information available, and unfortunately for me a lot of it is focused on modifiers of the C282Y genotype, rather than H63D. I started by looking at my genotype for each of the common modifier variants associated with iron overload in individuals with HH (helpfully reviewed by Rochette and colleagues). I also looked at the variants in TF and TMPRSS6 identified by Benyamin and colleagues as being associated with normal variation in measures of iron burden in a general population sample. Some of these variants aren’t included in my 23andMe data, so in these instances I used a SNP in very high linkage disequilibrium with the variant as a proxy. SNPs where I carry an allele associated with an elevated iron level are indicated in red.
Haemochromatosis modifiers associated with serum ferritin levels
| Gene | Variant | 23andMe SNP | My genotype [1] |
| TNF-alpha | rs1800629 | rs1800629 | GG |
| Mitochondria | 16189 | Haplogroup J [2] | T |
| CYBRD1 | rs884409 | rs3806563 | GG |
| BMP2 | rs235756 | rs235756 | GA |
Variants associated with normal variation in iron level markers
| Gene | Variant | 23andMe SNP | My genotype [1] | Trait |
| TF | rs3811647 | rs3811647 | GG | Serum transferrin |
| TMPRSS6 | rs855791 | rs855791 | GA | Serum iron |
[1] If the original report uses the negative strand I used the complement be consistent with 23andMe data. [2] I have haplogroup J which carries the T allele – thanks to Luke for working this out.
Overall these results aren’t very exciting; for most variants I do not have a genotype that is associated with higher iron levels. I did some further investigation of the genes where I am heterozygous for the variant (TMPRSS6 and BMP2) and found out a bit more about BMP2. Milet and colleagues identified an association between the rs235756 variant in BMP2, and variants in BMP4 and HJV, and pre-treatment ferritin levels in a clinical sample of individuals who were homozygous for the C282Y variant. My genotypes for these three variants are in the table below.
| Gene | Variant | 23andMe SNP | My genotype |
| BMP2 | rs235756 | rs235756 | CT [1] |
| BMP4 | rs4901474 | rs4901474 | CT |
| HJV | rs16827043 | rs16827043 | AA |
[1] I have used the complement alleles to be consistent with the figure below.
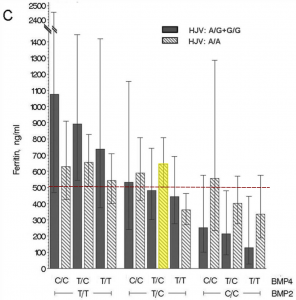
The relationship between these three variants and serum ferritin levels is best illustrated by the figure from their paper (below), in which I’ve coloured my genotype group in yellow and indicated the average ferritin level of the whole study sample by the dashed red line.
As you can see, individuals with my combination of genotypes at SNPs in BMP2, BMP4, and HJV had a higher-than-average mean ferritin level of about 650 ng/mL, adjusted for age and sex (the normal range for ferritin levels is about 12-300 ng/mL). The major caveat is that these findings only apply to individuals with the C282Y variant – they may not apply to people like me with H63D variants (another caveat is that a second study did not replicate these findings). Average ferritin levels are lower in H63D homozygotes than C282Y homozygotes (about 70 ng/mL in women and 130 ng/mL in men versus 145 ng/mL in women and 310 ng/mL in men respectively in a non-clinical sample).
What does this mean?
The short answer is: I’m not sure. I carry some variants that are associated with increased risk of iron overload, but I have no real idea of the effect of all these variants for someone homozygous for H63D. But supposing these variants do affect ferritin levels in people with H63D variants, it’s possible that my dad carries the risk alleles for the BMP2 and BMP4 variants (he could even have a copy of the HJV variant) and passed them on to me. This could go some way to explaining the differences between my family members, but in the absence of genotype data from everyone, I have no idea whether my conjecture matches reality.
So my attempt to find some genetic reason for why my dad and I have high ferritin levels has been interesting but essentially inconclusive. This is really due to lack of information, both in terms of the research that exists on this topic at the moment, and how much information I have about my genome and those of my family members. There might be a relatively common variant that interacts with H63D to increase risk of iron overload, I just have to wait for someone to find it. Or my dad and I could have some rare variant(s) that increase risk. Now I just need to convince my family to get their genomes sequenced…








 RSS
RSS Twitter
Twitter
at least you’re not brown. i’m kind of sick of 23andMe telling me i have lower risks for things i know i have higher risks for ;-)
Just a relatively irrelevant note: the mtDNA site 16189 is hypervariable and has mutated from T to C and back many many times in human history. I wouldn’t trust that disease association for a second – but I couldn’t be bothered to check the reference what they’re actually saying about it.
I wonder if there’s regulatory variation in these genes…
Hi,
You make quite some steps to convert your array results to family genetics. Some QC questions…
Has your sister’s test been done by sequencing or another test?
If it has been done by a locus-specific test (e.g. DNA oligo hybridisation based) it will probable miss the presence of a heterozygous deletion or local/nearby DNA variant that obscures the 2nd allel. Sequencing will miss the second allel when a disturbing SNP is within one of the PCR primers used for the generation of the amplicon to be sequenced. (or used for the oligohyb. test)
Has your test been confirmed by sequencing?
Most preferably would be to test DNA of your parents (or enough relevant relatives) to make sure that all parental alleles have been defined, before drawing these family genetics conclusions.
Hi,
There are no simple Mendelian disorders and Katherine provides an excellent example. There are children who have a genotype for PKU and carry high levels of phenylalanine but are phenotypically normal even thought they have not been treated with the appropriate diet. HH is probably more complex than many other disorders with its low penetrance, particularly with women.
@Tuuli
Yes, I agree with you about the mtDNA association. I’m not an expert on HH and I haven’t looked for eQTLs, but it seems entirely possible to me.
@Jasper
My sister had a clinical genetic test (in Australia). I’m can’t give you the specifics of how it was done but I don’t think it was sequencing. Ideally we’d all like to confirm our 23andMe results by sequencing but unfortunately the Genomes Unzipped budget doesn’t extend that far at the moment!
I was surprised to find out by 23andme that my husband and I are both carriers of the H63D hemochromatosis allele. I’m a regular blood donor, and so I never had (or probably wouldn’t have) any symptoms. We are fortunate that our only child is not even a carrier.
@ Katherine
I think 23andme should report a score per variant/probe of the intensity relative to a population and its spread (StDev?) in that population. Thus the local chromosome copy number.
In that manner it becomes more sure that a +/+ call is indeed a homozygous variant. That measure (i.e. local log2 sample/control ratio would indicate the presence of a Null allele and/or a local copy number variation (in this case a deletion).
That said, when incomplete penetrance comes into play for a certain genotype-fenotype, then your individual cloud of DNA variants (inherited and de novo) will separate you from your sister. Although one normaly can argue that you share part of the ‘other’ DNA variants.
This is an interesting story that likely will serve as an example for many that try to link genotype to phenotype for their personal data. It underscores the complexity that exists between genetic variation and phenotypic response.
You make mention in the replies of confirming the genotype data. That is good. I have questions of confirming your ferritin levels – How many times did you have ferritin assessed and how do those values change during a menstrual cycle? We’ve recognized that measures of certain hormones and cytokines important in adipogenesis and inflammation vary according with circadian rhythm. The importance of these naturally occurring rhythmic processes should not be overlooked when taking measures of clinically relevant molecules.
@Larry
Thanks for your comments – you make some good points. The next step in this personal genetics project is to go to my GP with my 23andMe results and get my ferritin levels tested again so watch this space!
this is very interesting because i was wondering about all of that my father passed away not knowing he had the disease until recently my sister got really sick about 4yrs ago and found out she had hemochromotosis cy282 and the h63d so i have it to both those and i just found that my ten yr old has it to but he has the two h63d mutated genes but my daughter only has one so what does it mean because he is also got a low rdw and a high eosinophil so when i was reading it means something like he is anemic. See my mother is like that she has one gene and is anemic so i really am looking into this