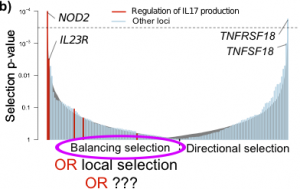
 As I mentioned a few weeks ago, we recently published a large study into the genetics of inflammatory bowel disease (IBD), which included a number of analyses digging into the biology and evolutionary history of IBD genetic risk. Gratifyingly, our paper has stimulated a lot of discussion among other scientists, which has generated several ideas about future directions for this work. One question that was raised by several population-genetics experts at ASHG was about our natural selection analysis, and in particular our claim to discover an enrichment of balancing selection in IBD loci. In the paper, we found clear signals of natural selection on IBD loci, a subset of which we interpreted as balancing selection. In this post I will set out how I came to this conclusion, but then outline another explanation that could explain the results: recent local positive selection in Europeans.
As I mentioned a few weeks ago, we recently published a large study into the genetics of inflammatory bowel disease (IBD), which included a number of analyses digging into the biology and evolutionary history of IBD genetic risk. Gratifyingly, our paper has stimulated a lot of discussion among other scientists, which has generated several ideas about future directions for this work. One question that was raised by several population-genetics experts at ASHG was about our natural selection analysis, and in particular our claim to discover an enrichment of balancing selection in IBD loci. In the paper, we found clear signals of natural selection on IBD loci, a subset of which we interpreted as balancing selection. In this post I will set out how I came to this conclusion, but then outline another explanation that could explain the results: recent local positive selection in Europeans.
Continue reading ‘Looking closer at natural selection in inflammatory bowel disease’
 RSS
RSS Twitter
Twitter
Recent Comments