 This is a guest post by Peter Cheng and Eliana Hechter from the University of California, Berkeley.
This is a guest post by Peter Cheng and Eliana Hechter from the University of California, Berkeley.
Suppose that you’ve had your DNA genotyped by 23andMe or some other DTC genetic testing company. Then an article shows up in your morning newspaper or journal (like this one) and suddenly there’s an additional variant you want to know about. You check your raw genotypes file to see if the variant is present on the chip, but it isn’t! So what next? [Note: the most recent 23andMe chip does include this variant, although older versions of their chip do not.]
Genotype imputation is a process used for predicting, or “imputing”, genotypes that are not assayed by a genotyping chip. The process compares the genotyped data from a chip (e.g. your 23andMe results) with a reference panel of genomes (supplied by big genome projects like the 1000 Genomes or HapMap projects) in order to make predictions about variants that aren’t on the chip. If you want a technical review of imputation (and the program IMPUTE in particular), we recommend Marchini & Howie’s 2010 Nature Reviews Genetics article. However, the following figure provides an intuitive understanding of the process.
Continue reading ‘Learning more from your 23andMe results with Imputation’
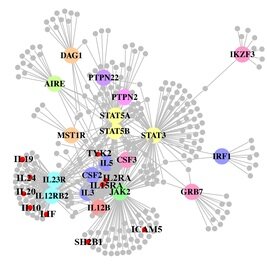 A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ]
A paper out in PLoS Genetics this week takes a step towards using genome-wide association data to reconstruct functional pathways. Using protein-protein interaction data and tissue-specific expression data, the authors reconstruct biochemical pathways that underlie various diseases, by looking for variants that interact with genes in GWAS regions. These networks can then tell us about what systems are disrupted by GWAS variants as a whole, as well as identifying potential drug targets. The figure to the right shows the network constructed for Crohn’s disease; large colored circles are genes in GWAS loci, small grey circles are other genes in the network they constructed. As an interesting side note, the GWAS variants were taken from a 2008 study; since then, we have published a new meta-analysis, which implicated a lot of new regions. 10 genes in these regions, marked as small red circles on the figure, were also in the disease network. [LJ] RSS
RSS Twitter
Twitter
Recent Comments